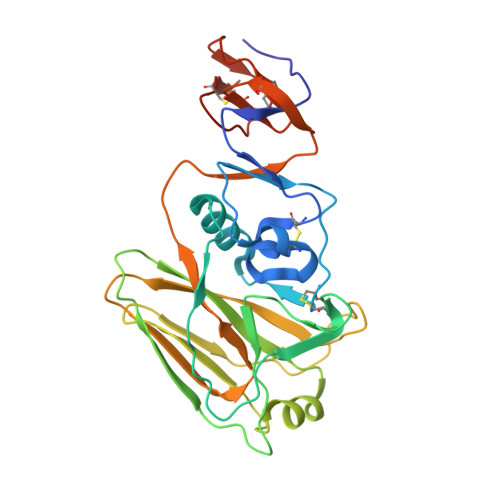
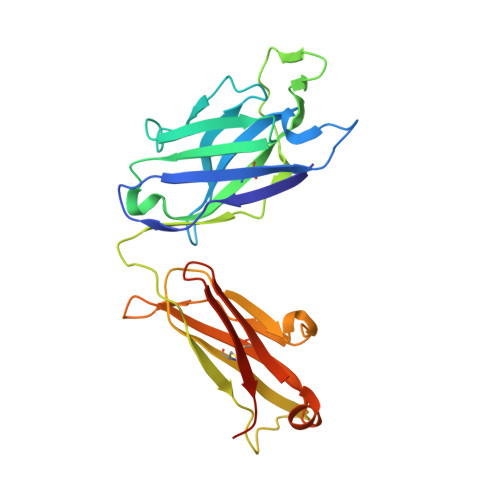
Multistate design of influenza antibodies improves affinity and breadth against seasonal viruses.
Sevy, A.M., Wu, N.C., Gilchuk, I.M., Parrish, E.H., Burger, S., Yousif, D., Nagel, M.B.M., Schey, K.L., Wilson, I.A., Crowe Jr., J.E., Meiler, J.(2019) Proc Natl Acad Sci U S A 116: 1597-1602
- PubMed: 30642961
- DOI: https://doi.org/10.1073/pnas.1806004116
- Primary Citation of Related Structures:
6D0U - PubMed Abstract:
Influenza is a yearly threat to global public health. Rapid changes in influenza surface proteins resulting from antigenic drift and shift events make it difficult to readily identify antibodies with broadly neutralizing activity against different influenza subtypes with high frequency, specifically antibodies targeting the receptor binding domain (RBD) on influenza HA protein. We developed an optimized computational design method that is able to optimize an antibody for recognition of large panels of antigens. To demonstrate the utility of this multistate design method, we used it to redesign an antiinfluenza antibody against a large panel of more than 500 seasonal HA antigens of the H1 subtype. As a proof of concept, we tested this method on a variety of known antiinfluenza antibodies and identified those that could be improved computationally. We generated redesigned variants of antibody C05 to the HA RBD and experimentally characterized variants that exhibited improved breadth and affinity against our panel. C05 mutants exhibited improved affinity for three of the subtypes used in design by stabilizing the CDRH3 loop and creating favorable electrostatic interactions with the antigen. These mutants possess increased breadth and affinity of binding while maintaining high-affinity binding to existing targets, surpassing a major limitation up to this point.
- Chemical & Physical Biology Program, Vanderbilt University, Nashville, TN 37235.
Organizational Affiliation: