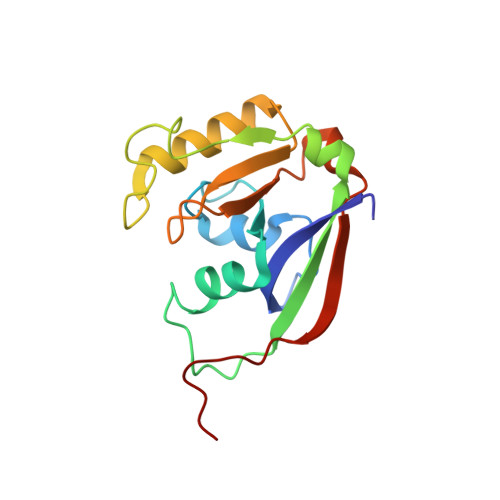
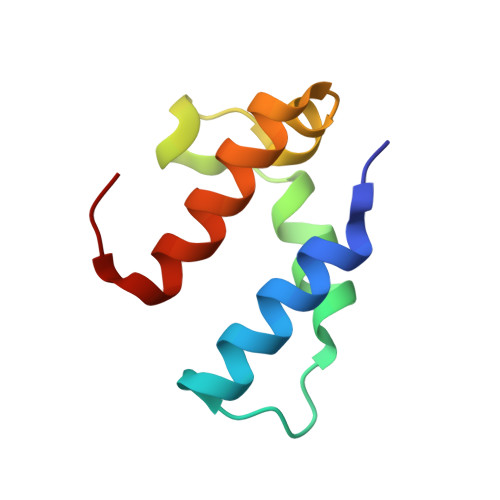
ParST is a widespread toxin-antitoxin module that targets nucleotide metabolism.
Piscotta, F.J., Jeffrey, P.D., Link, A.J.(2019) Proc Natl Acad Sci U S A 116: 826-834
- PubMed: 30598453
- DOI: https://doi.org/10.1073/pnas.1814633116
- Primary Citation of Related Structures:
6D0H, 6D0I - PubMed Abstract:
Toxin-antitoxin (TA) systems interfere with essential cellular processes and are implicated in bacterial lifestyle adaptations such as persistence and the biofilm formation. Here, we present structural, biochemical, and functional data on an uncharacterized TA system, the COG5654-COG5642 pair. Bioinformatic analysis showed that this TA pair is found in 2,942 of the 16,286 distinct bacterial species in the RefSeq database. We solved a structure of the toxin bound to a fragment of the antitoxin to 1.50 Å. This structure suggested that the toxin is a mono-ADP-ribosyltransferase (mART). The toxin specifically modifies phosphoribosyl pyrophosphate synthetase (Prs), an essential enzyme in nucleotide biosynthesis conserved in all organisms. We propose renaming the toxin ParT for Prs ADP-ribosylating toxin and ParS for the cognate antitoxin. ParT is a unique example of an intracellular protein mART in bacteria and is the smallest known mART. This work demonstrates that TA systems can induce bacteriostasis through interference with nucleotide biosynthesis.
- Department of Chemical and Biological Engineering, Princeton University, Princeton, NJ 08544.
Organizational Affiliation: