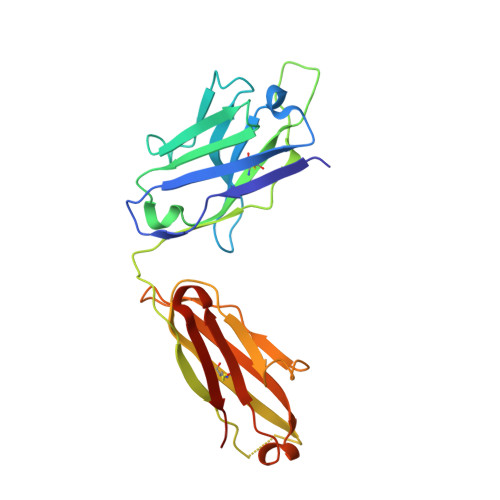
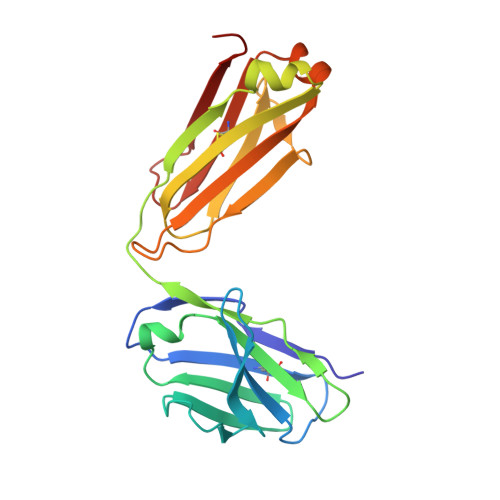
Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope.
Imkeller, K., Scally, S.W., Bosch, A., Marti, G.P., Costa, G., Triller, G., Murugan, R., Renna, V., Jumaa, H., Kremsner, P.G., Sim, B.K.L., Hoffman, S.L., Mordmuller, B., Levashina, E.A., Julien, J.P., Wardemann, H.(2018) Science 360: 1358-1362
- PubMed: 29880723
- DOI: https://doi.org/10.1126/science.aar5304
- Primary Citation of Related Structures:
6D01, 6D0X, 6D11 - PubMed Abstract:
Affinity maturation selects B cells expressing somatically mutated antibody variants with improved antigen-binding properties to protect from invading pathogens. We determined the molecular mechanism underlying the clonal selection and affinity maturation of human B cells expressing protective antibodies against the circumsporozoite protein of the malaria parasite Plasmodium falciparum (PfCSP). We show in molecular detail that the repetitive nature of PfCSP facilitates direct homotypic interactions between two PfCSP repeat-bound monoclonal antibodies, thereby improving antigen affinity and B cell activation. These data provide a mechanistic explanation for the strong selection of somatic mutations that mediate homotypic antibody interactions after repeated parasite exposure in humans. Our findings demonstrate a different mode of antigen-mediated affinity maturation to improve antibody responses to PfCSP and presumably other repetitive antigens.
- B Cell Immunology, German Cancer Research Institute, Heidelberg, Germany.
Organizational Affiliation: