Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Ma, J., Lei, H.T., Reyes, F.E., Sanchez-Martinez, S., Sarhan, M.F., Hattne, J., Gonen, T.(2019) Proc Natl Acad Sci U S A 116: 2086-2090
- PubMed: 30659158
- DOI: https://doi.org/10.1073/pnas.1806206116
- Primary Citation of Related Structures:
6CSE, 6CSF - PubMed Abstract:
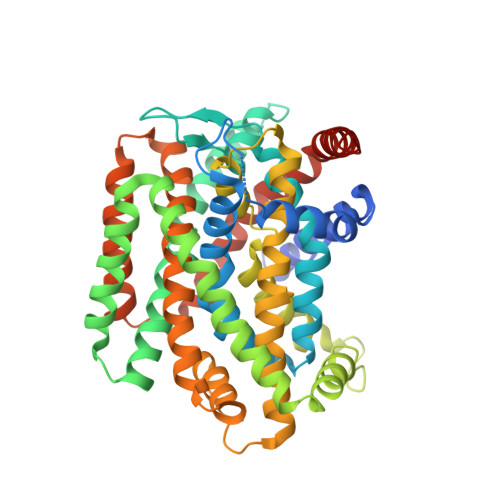
The amino acid, polyamine, and organocation (APC) superfamily is the second largest superfamily of membrane proteins forming secondary transporters that move a range of organic molecules across the cell membrane. Each transporter in the APC superfamily is specific for a unique subset of substrates, even if they possess a similar structural fold. The mechanism of substrate selectivity remains, by and large, elusive. Here, we report two crystal structures of an APC member from Methanococcus maripaludis , the alanine or glycine:cation symporter (AgcS), with l- or d-alanine bound. Structural analysis combined with site-directed mutagenesis and functional studies inform on substrate binding, specificity, and modulation of the AgcS family and reveal key structural features that allow this transporter to accommodate glycine and alanine while excluding all other amino acids. Mutation of key residues in the substrate binding site expand the selectivity to include valine and leucine. These studies provide initial insights into substrate selectivity in AgcS symporters.
- Janelia Research Campus, Howard Hughes Medical Institute, Ashburn, VA 20147.
Organizational Affiliation: