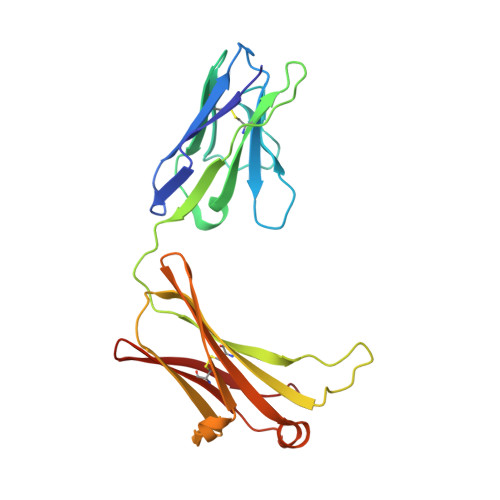
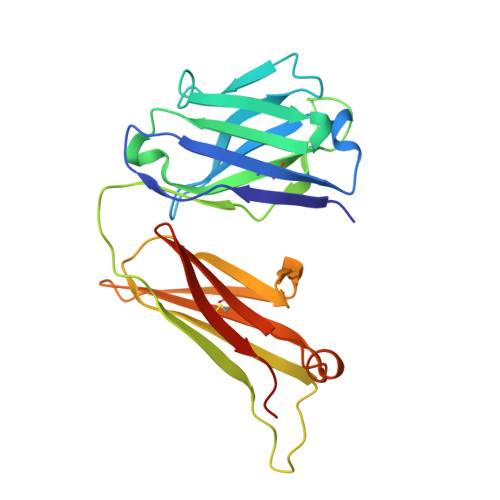
EFab domain substitution as a solution to the light-chain pairing problem of bispecific antibodies.
Cooke, H.A., Arndt, J., Quan, C., Shapiro, R.I., Wen, D., Foley, S., Vecchi, M.M., Preyer, M.(2018) MAbs 10: 1248-1259
- PubMed: 30215570
- DOI: https://doi.org/10.1080/19420862.2018.1519631
- Primary Citation of Related Structures:
6CR1 - PubMed Abstract:
Bispecific antibody therapeutics can expand the functionality of a conventional monoclonal antibody drug because they can bind multiple antigens. However, their great potential is counterbalanced by the challenges faced in their production. The classic asymmetric bispecific containing an Fc requires the expression of four unique chains - two light chains and two heavy chains; each light chain must pair with its correct heavy chain, which then must heterodimerize to form the full bispecific. The light-chain pairing problem has several solutions, some of which require engineering and optimization for each bispecific pair. Here, we introduce a technology called EFab Domain Substitution, which replaces the Cε2 of IgE for one of the CL/CH1 domains into one arm of an asymmetric bispecific to encourage the correct pairing of the light chains. EFab Domain Substitution provides very robust correct pairing while maintaining antibody function and is effective for many variable domains. We report its effect on the biophysical properties of an antibody and the crystal structure of the EFab domain substituted into the adalimumab Fab (PDB ID 6CR1).
- a Department of Biotherapeutic and Medicinal Sciences , Biogen , Cambridge , MA , USA.
Organizational Affiliation: