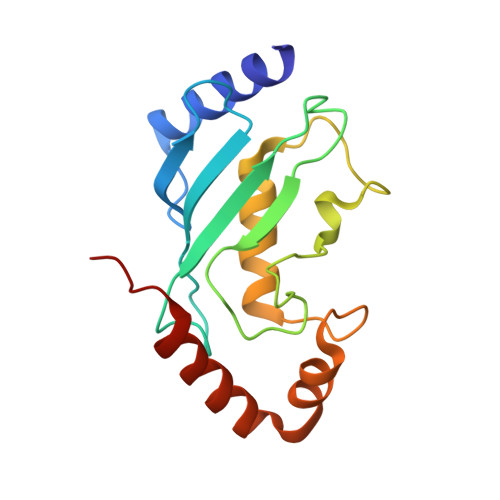
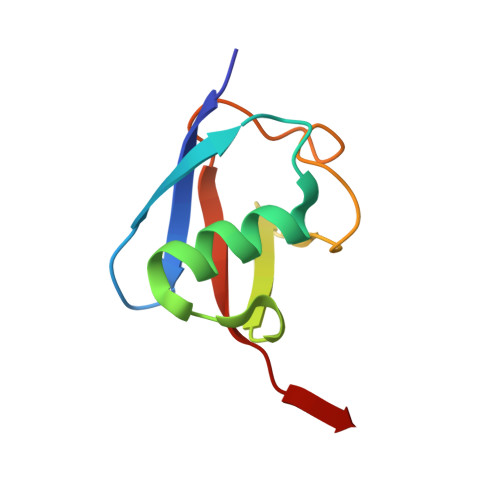
Insights into the ubiquitin transfer cascade catalyzed by theLegionellaeffector SidC.
Wasilko, D.J., Huang, Q., Mao, Y.(2018) Elife 7
- PubMed: 30015617
- DOI: https://doi.org/10.7554/eLife.36154
- Primary Citation of Related Structures:
6CP0, 6CP2 - PubMed Abstract:
The causative agent of Legionnaires' disease, Legionella pneumophila , delivers more than 330 virulent effectors to its host to establish an intracellular membrane-bound organelle called the Legionella containing vacuole. Among the army of Legionella effectors, SidC and its paralog SdcA have been identified as novel bacterial ubiquitin (Ub) E3 ligases. To gain insight into the molecular mechanism of SidC/SdcA as Ub ligases, we determined the crystal structures of a binary complex of the N-terminal catalytic SNL domain of SdcA with its cognate E2 UbcH5C and a ternary complex consisting of the SNL domain of SidC with the Ub-linked E2 UbcH7. These two structures reveal the molecular determinants governing the Ub transfer cascade catalyzed by SidC. Together, our data support a common mechanism in the Ub transfer cascade in which the donor Ub is immobilized with its C-terminal tail locked in an extended conformation, priming the donor Ub for catalysis.
- Department of Molecular Biology and Genetics, Cornell University, Ithaca, United States.
Organizational Affiliation: