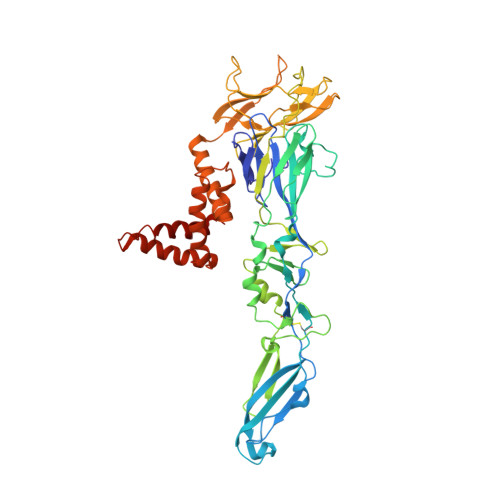
Refinement and Analysis of the Mature Zika Virus Cryo-EM Structure at 3.1 angstrom Resolution.
Sevvana, M., Long, F., Miller, A.S., Klose, T., Buda, G., Sun, L., Kuhn, R.J., Rossmann, M.G.(2018) Structure 26: 1169
- PubMed: 29958768
- DOI: https://doi.org/10.1016/j.str.2018.05.006
- Primary Citation of Related Structures:
6CO8 - PubMed Abstract:
Among the several arthropod-borne human flaviviral diseases, the recent outbreak of Zika virus (ZIKV) has caused devastating birth defects and neurological disorders, challenging the world with another major public health concern. We report here the refined structure of the mature ZIKV at a resolution of 3.1 Å as determined by cryo-electron microscopic single-particle reconstruction. The improvement in the resolution, compared with previous enveloped virus structures, was the result of optimized virus preparation methods and data processing techniques. The glycoprotein interactions and surface properties of ZIKV were compared with other mosquito-borne flavivirus structures. The largest structural differences and sequence variations occur at the glycosylation loop associated with receptor binding. Probable drug binding pockets were identified on the viral surface. These results also provide a structural basis for the design of vaccines against ZIKV.
- Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA.
Organizational Affiliation: