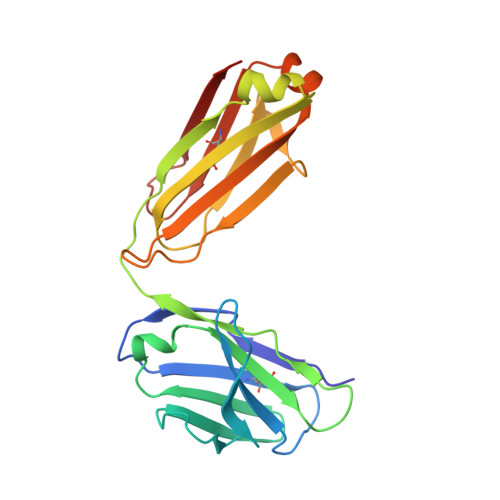
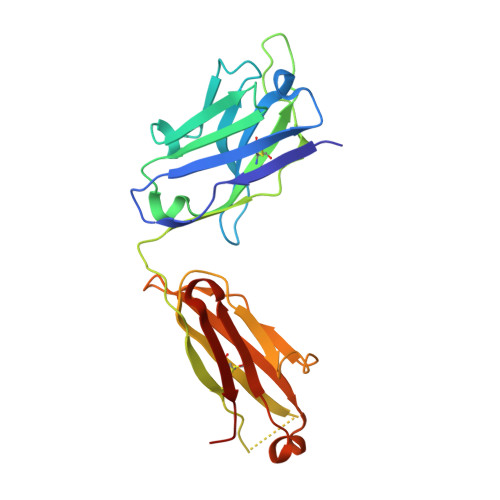
Structural and kinetic basis for the selectivity of aducanumab for aggregated forms of amyloid-beta.
Arndt, J.W., Qian, F., Smith, B.A., Quan, C., Kilambi, K.P., Bush, M.W., Walz, T., Pepinsky, R.B., Bussiere, T., Hamann, S., Cameron, T.O., Weinreb, P.H.(2018) Sci Rep 8: 6412-6412
- PubMed: 29686315
- DOI: https://doi.org/10.1038/s41598-018-24501-0
- Primary Citation of Related Structures:
6CNR, 6CO3 - PubMed Abstract:
Aducanumab, a human-derived antibody targeting amyloid-β (Aβ), is in Phase 3 clinical trials for the treatment of Alzheimer's disease. Biochemical and structural analyses show that aducanumab binds a linear epitope formed by amino acids 3-7 of the Aβ peptide. Aducanumab discriminates between monomers and oligomeric or fibrillar aggregates based on weak monovalent affinity, fast binding kinetics and strong avidity for epitope-rich aggregates. Direct comparative studies with analogs of gantenerumab, bapineuzumab and solanezumab demonstrate clear differentiation in the binding properties of these antibodies. The crystal structure of the Fab fragment of aducanumab bound to its epitope peptide reveals that aducanumab binds to the N terminus of Aβ in an extended conformation, distinct from those seen in structures with other antibodies that target this immunodominant epitope. Aducanumab recognizes a compact epitope that sits in a shallow pocket on the antibody surface. In silico analyses suggest that aducanumab interacts weakly with the Aβ monomer and may accommodate a variety of peptide conformations, further supporting its selectivity for Aβ aggregates. Our studies provide a structural rationale for the low affinity of aducanumab for non-pathogenic monomers and its greater selectivity for aggregated forms than is seen for other Aβ-targeting antibodies.
- Biogen, Cambridge, MA, USA. joe.arndt@biogen.com.
Organizational Affiliation: