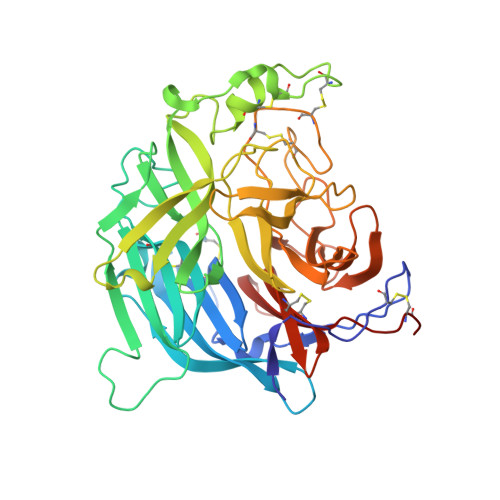
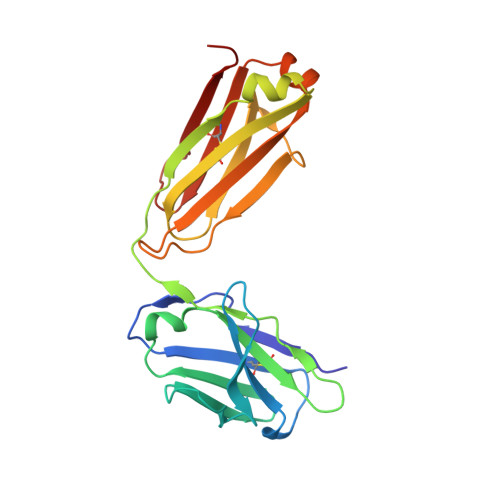
Crystal structure of the Hendra virus attachment G glycoprotein bound to a potent cross-reactive neutralizing human monoclonal antibody.
Xu, K., Rockx, B., Xie, Y., DeBuysscher, B.L., Fusco, D.L., Zhu, Z., Chan, Y.P., Xu, Y., Luu, T., Cer, R.Z., Feldmann, H., Mokashi, V., Dimitrov, D.S., Bishop-Lilly, K.A., Broder, C.C., Nikolov, D.B.(2013) PLoS Pathog 9: e1003684
- PubMed: 24130486
- DOI: https://doi.org/10.1371/journal.ppat.1003684
- Primary Citation of Related Structures:
6CMG, 6CMI - PubMed Abstract:
The henipaviruses, represented by Hendra (HeV) and Nipah (NiV) viruses are highly pathogenic zoonotic paramyxoviruses with uniquely broad host tropisms responsible for repeated outbreaks in Australia, Southeast Asia, India and Bangladesh. The high morbidity and mortality rates associated with infection and lack of licensed antiviral therapies make the henipaviruses a potential biological threat to humans and livestock. Henipavirus entry is initiated by the attachment of the G envelope glycoprotein to host cell membrane receptors. Previously, henipavirus-neutralizing human monoclonal antibodies (hmAb) have been isolated using the HeV-G glycoprotein and a human naïve antibody library. One cross-reactive and receptor-blocking hmAb (m102.4) was recently demonstrated to be an effective post-exposure therapy in two animal models of NiV and HeV infection, has been used in several people on a compassionate use basis, and is currently in development for use in humans. Here, we report the crystal structure of the complex of HeV-G with m102.3, an m102.4 derivative, and describe NiV and HeV escape mutants. This structure provides detailed insight into the mechanism of HeV and NiV neutralization by m102.4, and serves as a blueprint for further optimization of m102.4 as a therapeutic agent and for the development of entry inhibitors and vaccines.
- Structural Biology Program, Memorial Sloan Kettering Cancer Center, New York, New York, United States of America.
Organizational Affiliation: