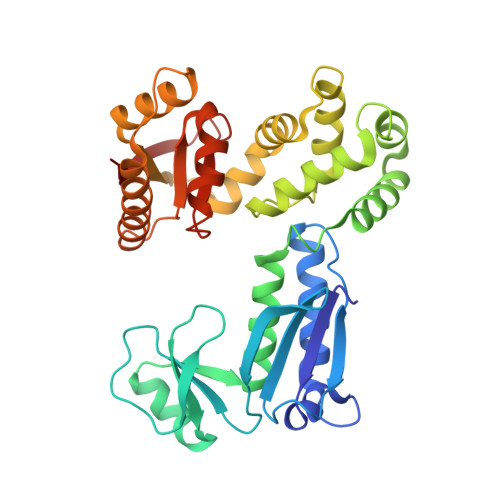
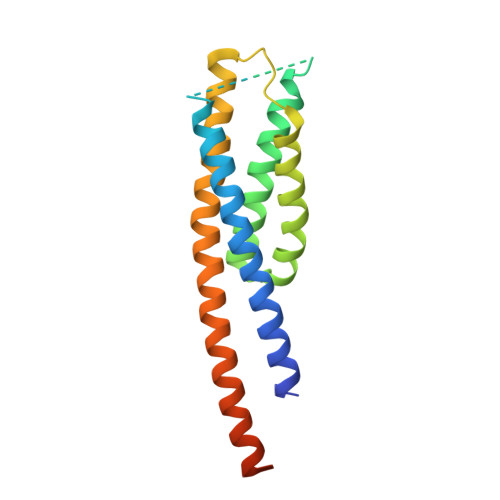
Structures of chaperone-substrate complexes docked onto the export gate in a type III secretion system.
Xing, Q., Shi, K., Portaliou, A., Rossi, P., Economou, A., Kalodimos, C.G.(2018) Nat Commun 9: 1773-1773
- PubMed: 29720631
- DOI: https://doi.org/10.1038/s41467-018-04137-4
- Primary Citation of Related Structures:
6CH1, 6CH2, 6CH3 - PubMed Abstract:
The flagellum and the injectisome enable bacterial locomotion and pathogenesis, respectively. These nanomachines assemble and function using a type III secretion system (T3SS). Exported proteins are delivered to the export apparatus by dedicated cytoplasmic chaperones for their transport through the membrane. The structural and mechanistic basis of this process is poorly understood. Here we report the structures of two ternary complexes among flagellar chaperones (FliT and FliS), protein substrates (the filament-capping FliD and flagellin FliC), and the export gate platform protein FlhA. The substrates do not interact directly with FlhA; however, they are required to induce a binding-competent conformation to the chaperone that exposes the recognition motif featuring a highly conserved sequence recognized by FlhA. The structural data reveal the recognition signal in a class of T3SS proteins and provide new insight into the assembly of key protein complexes at the export gate.
- Department of Structural Biology, St. Jude Children's Research Hospital, 263 Danny Thomas Place, Memphis, TN, 38105, USA.
Organizational Affiliation: