Revised Crystal Structure of Human Adenovirus Reveals the Limits on Protein IX Quasi-Equivalence and on Analyzing Large Macromolecular Complexes.
Kundhavai Natchiar, S., Venkataraman, S., Mullen, T.M., Nemerow, G.R., Reddy, V.S.(2018) J Mol Biology 430: 4132-4141
- PubMed: 30121295
- DOI: https://doi.org/10.1016/j.jmb.2018.08.011
- Primary Citation of Related Structures:
6CGV - PubMed Abstract:
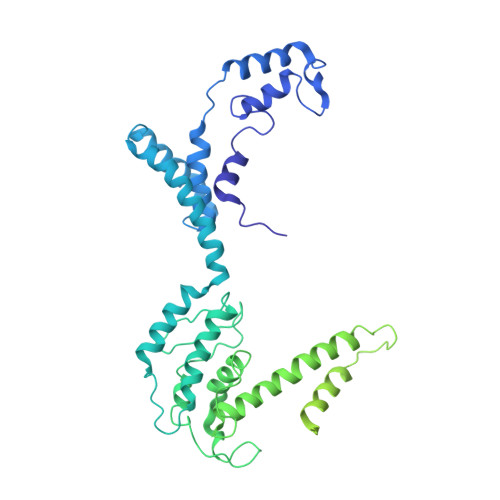
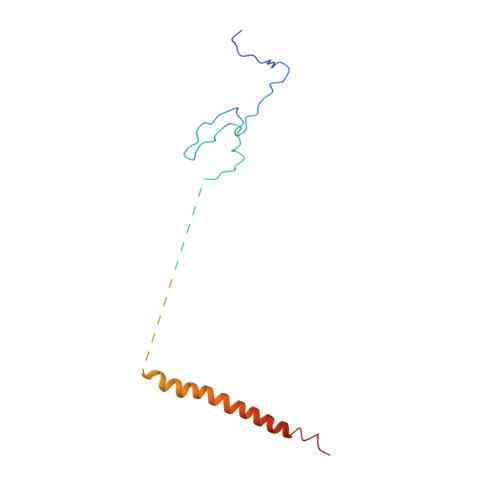
We report the revised crystal structure of a pseudo-typed human adenovirus at 3.8-Å resolution that is consistent with the atomic models of minor proteins determined by cryo-electron microscopy. The diffraction data from multiple crystals were rescaled and merged to increase the data completeness. The densities for the minor proteins were initially identified in the phase-refined omit maps that were further improved by the phases from docked poly-alanine models to build atomic structures. While the trimeric fiber molecules are disordered due to flexibility and imposition of 5-fold symmetry, the remaining major capsid proteins hexon and penton base are clearly ordered, with the exception of hypervariable region 1 of hexons, the RGD containing loop, and the N-termini of the penton base. The exterior minor protein IX together with the interior minor proteins IIIa and VIII stabilizes the adenovirus virion. A segment of N-terminal pro-peptide of VI is found in the interior cavities of peripentonal hexons, and the rest of VI is disordered. While the triskelion substructures formed by the N-termini of IX conform to excellent quasi 3-fold symmetry, the tetrameric coiled-coils formed by the C-termini and organized in parallel and anti-parallel arrangement do not exhibit any quasi-symmetry. This observation also conveys the pitfalls of using the quasi-equivalence as validation criteria for the structural analysis of extended (non-modular) capsid proteins such as IX. Together, these results remedy certain discrepancies in the previous X-ray model in agreement with the cryo-electron microscopy models.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: