Rational Design of a 310-Helical PIP-Box Mimetic Targeting PCNA, the Human Sliding Clamp.
Wegener, K.L., McGrath, A.E., Dixon, N.E., Oakley, A.J., Scanlon, D.B., Abell, A.D., Bruning, J.B.(2018) Chemistry 24: 11325-11331
- PubMed: 29917264
- DOI: https://doi.org/10.1002/chem.201801734
- Primary Citation of Related Structures:
6CBI, 6CEJ, 6CIV, 6CIX - PubMed Abstract:
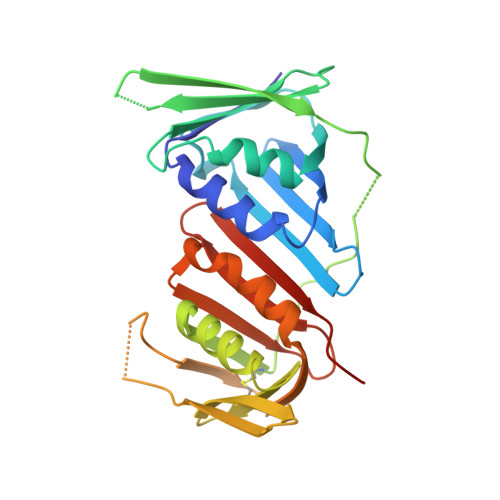
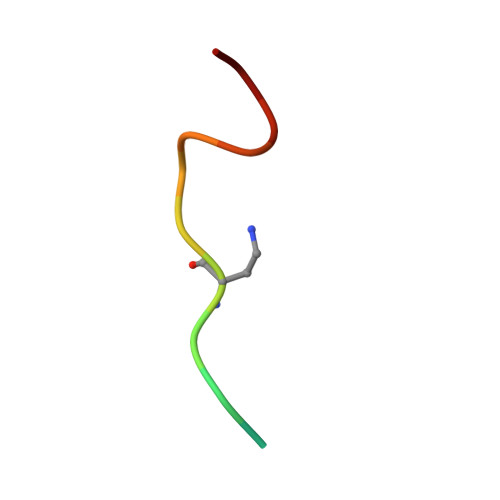
The human sliding clamp (PCNA) controls access to DNA for many proteins involved in DNA replication and repair. Proteins are recruited to the PCNA surface by means of a short, conserved peptide motif known as the PCNA-interacting protein box (PIP-box). Inhibitors of these essential protein-protein interactions may be useful as cancer therapeutics by disrupting DNA replication and repair in these highly proliferative cells. PIP-box peptide mimetics have been identified as a potentially rapid route to potent PCNA inhibitors. Here we describe the rational design and synthesis of the first PCNA peptidomimetic ligands, based on the high affinity PIP-box sequence from the natural PCNA inhibitor p21. These mimetics incorporate covalent i,i+4 side-chain/side-chain lactam linkages of different lengths, designed to constrain the peptides into the 3 10 -helical structure required for PCNA binding. NMR studies confirmed that while the unmodified p21 peptide had little defined structure in solution, mimetic ACR2 pre-organized into 3 10 -helical structure prior to interaction with PCNA. ACR2 displayed higher affinity binding than most known PIP-box peptides, and retains the native PCNA binding mode, as observed in the co-crystal structure of ACR2 bound to PCNA. This study offers a promising new strategy for PCNA inhibitor design for use as anti-cancer therapeutics.
- Institute for Photonics and Advanced Sensing (IPAS), School of Biological Sciences, The University of Adelaide, South Australia, 5005, Australia.
Organizational Affiliation: