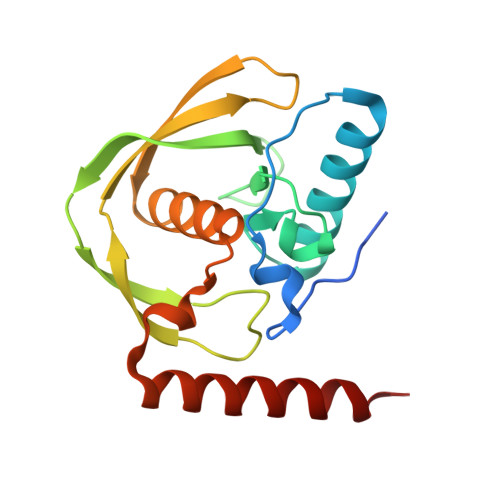
Structures of Legionella pneumophila serogroup 1 peptide deformylase bound to nickel(II) and actinonin
Nguyen, C.L., Fan, W., Fisher, S., Matthews, K., Norman, J.O., Abendroth, J., Barrett, K.F., Craig, J.K., Edwards, T.E., Lorimer, D.D., McLaughlin, K.J.(2025) Acta Crystallogr F Struct Biol Commun 81: 163-170