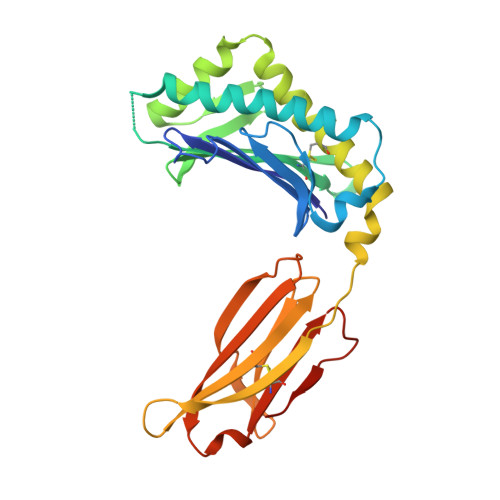
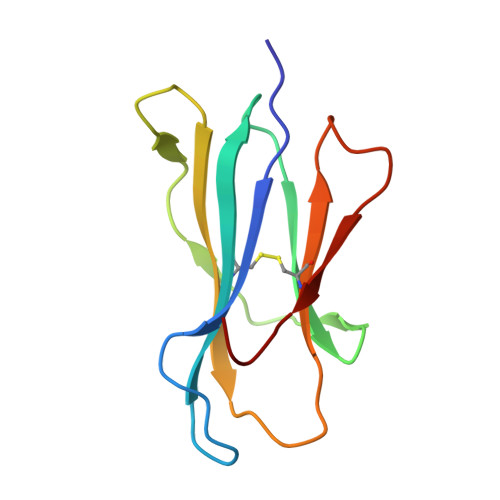
Insight into small molecule binding to the neonatal Fc receptor by X-ray crystallography and 100 kHz magic-angle-spinning NMR.
Stoppler, D., Macpherson, A., Smith-Penzel, S., Basse, N., Lecomte, F., Deboves, H., Taylor, R.D., Norman, T., Porter, J., Waters, L.C., Westwood, M., Cossins, B., Cain, K., White, J., Griffin, R., Prosser, C., Kelm, S., Sullivan, A.H., Fox, D., Carr, M.D., Henry, A., Taylor, R., Meier, B.H., Oschkinat, H., Lawson, A.D.(2018) PLoS Biol 16: e2006192-e2006192
- PubMed: 29782488
- DOI: https://doi.org/10.1371/journal.pbio.2006192
- Primary Citation of Related Structures:
6C97, 6C98, 6C99 - PubMed Abstract:
Aiming at the design of an allosteric modulator of the neonatal Fc receptor (FcRn)-Immunoglobulin G (IgG) interaction, we developed a new methodology including NMR fragment screening, X-ray crystallography, and magic-angle-spinning (MAS) NMR at 100 kHz after sedimentation, exploiting very fast spinning of the nondeuterated soluble 42 kDa receptor construct to obtain resolved proton-detected 2D and 3D NMR spectra. FcRn plays a crucial role in regulation of IgG and serum albumin catabolism. It is a clinically validated drug target for the treatment of autoimmune diseases caused by pathogenic antibodies via the inhibition of its interaction with IgG. We herein present the discovery of a small molecule that binds into a conserved cavity of the heterodimeric, extracellular domain composed of an α-chain and β2-microglobulin (β2m) (FcRnECD, 373 residues). X-ray crystallography was used alongside NMR at 100 kHz MAS with sedimented soluble protein to explore possibilities for refining the compound as an allosteric modulator. Proton-detected MAS NMR experiments on fully protonated [13C,15N]-labeled FcRnECD yielded ligand-induced chemical-shift perturbations (CSPs) for residues in the binding pocket and allosteric changes close to the interface of the two receptor heterodimers present in the asymmetric unit as well as potentially in the albumin interaction site. X-ray structures with and without ligand suggest the need for an optimized ligand to displace the α-chain with respect to β2m, both of which participate in the FcRnECD-IgG interaction site. Our investigation establishes a method to characterize structurally small molecule binding to nondeuterated large proteins by NMR, even in their glycosylated form, which may prove highly valuable for structure-based drug discovery campaigns.
- Leibniz-Forschungsinstitut für Molekulare Pharmakologie, Berlin, Germany.
Organizational Affiliation: