Evidence for two-tiered conformation selection in the SAM-III riboswitch
Price, I.R., Grigg, J.C., Martell, D.J., Ding, F., Peng, C., Ke, A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| SAM-III riboswitch | 47 | Enterococcus faecalis | 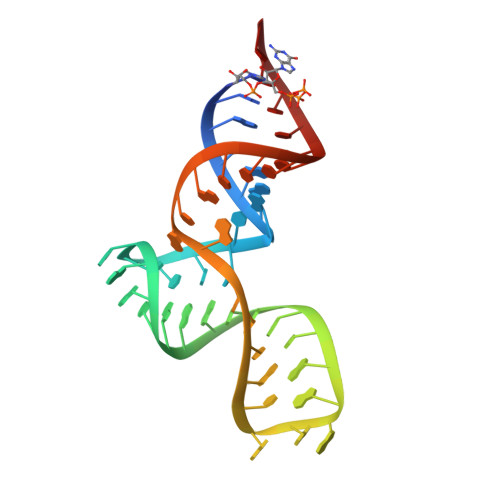 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NCO Query on NCO | C [auth A] D [auth A] E [auth A] F [auth A] G [auth B] | COBALT HEXAMMINE(III) Co H18 N6 DYLMFCCYOUSRTK-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 74.011 | α = 90 |
| b = 96.791 | β = 90 |
| c = 168.882 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data scaling |
| SOLVE | phasing |
| PDB_EXTRACT | data extraction |
| DENZO | data reduction |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM-086766 |