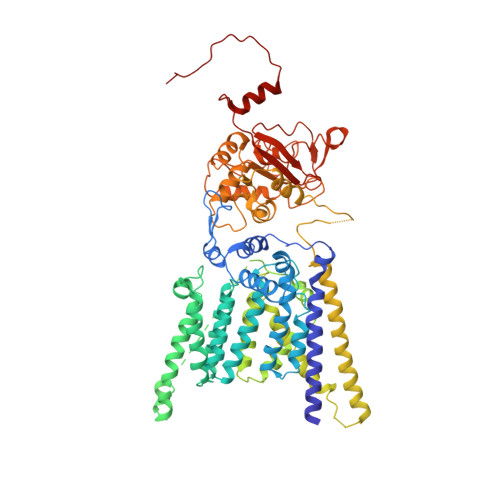
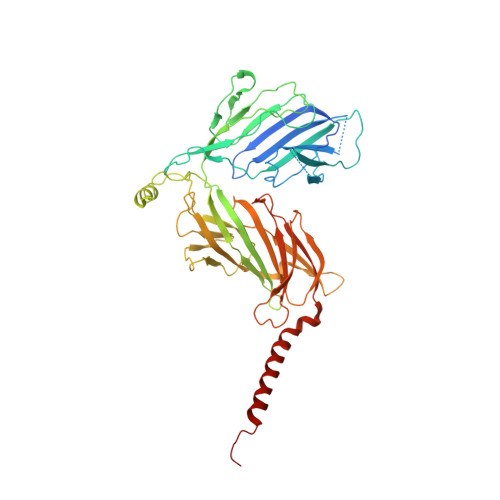
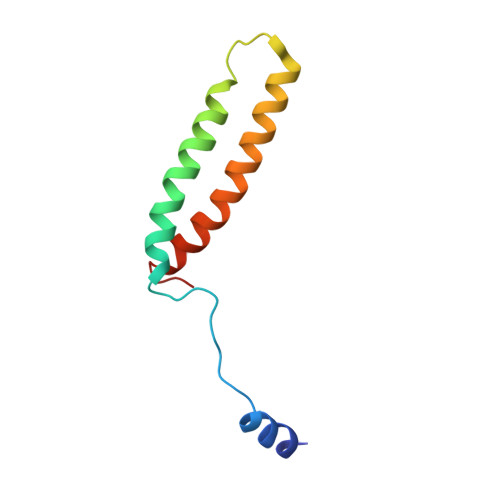
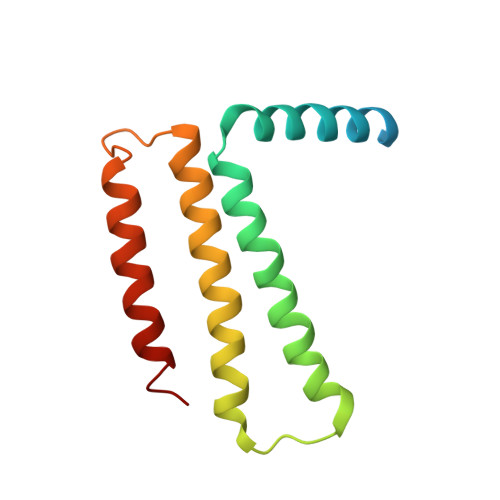
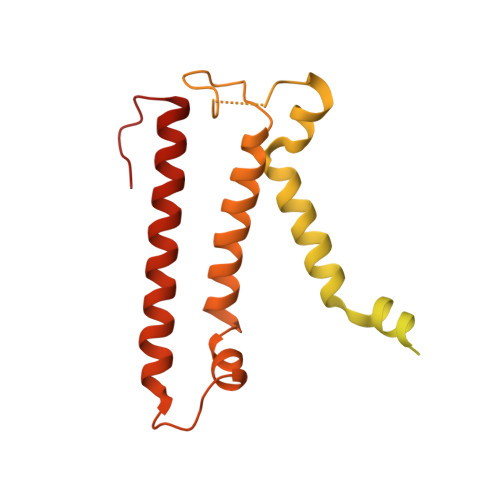
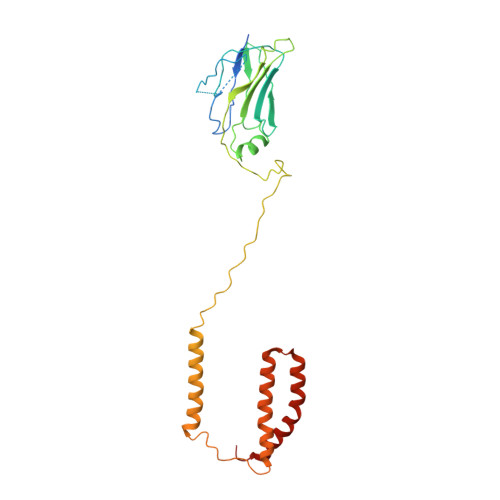
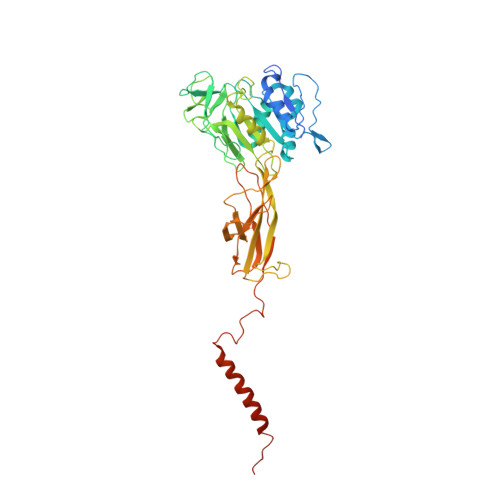
The atomic structure of a eukaryotic oligosaccharyltransferase complex.
Bai, L., Wang, T., Zhao, G., Kovach, A., Li, H.(2018) Nature 555: 328-333
- PubMed: 29466327
- DOI: https://doi.org/10.1038/nature25755
- Primary Citation of Related Structures:
6C26 - PubMed Abstract:
N-glycosylation is a ubiquitous modification of eukaryotic secretory and membrane-bound proteins; about 90% of glycoproteins are N-glycosylated. The reaction is catalysed by an eight-protein oligosaccharyltransferase (OST) complex that is embedded in the endoplasmic reticulum membrane. Our understanding of eukaryotic protein N-glycosylation has been limited owing to the lack of high-resolution structures. Here we report a 3.5 Å resolution cryo-electron microscopy structure of the Saccharomyces cerevisiae OST complex, revealing the structures of subunits Ost1-Ost5, Stt3, Wbp1 and Swp1. We found that seven phospholipids mediate many of the inter-subunit interactions, and an Stt3 N-glycan mediates interactions with Wbp1 and Swp1 in the lumen. Ost3 was found to mediate the OST-Sec61 translocon interface, funnelling the acceptor peptide towards the OST catalytic site as the nascent peptide emerges from the translocon. The structure provides insights into co-translational protein N-glycosylation, and may facilitate the development of small-molecule inhibitors that target this process.
- Center for Epigenetics, Van Andel Research Institute, Grand Rapids, Michigan, USA.
Organizational Affiliation: