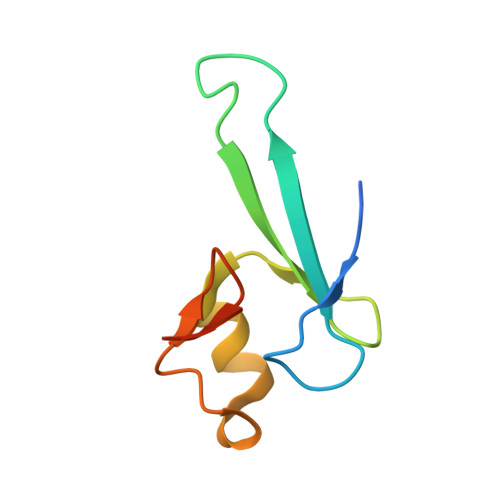
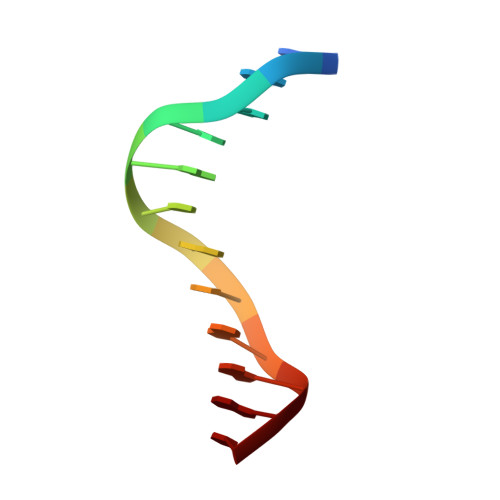
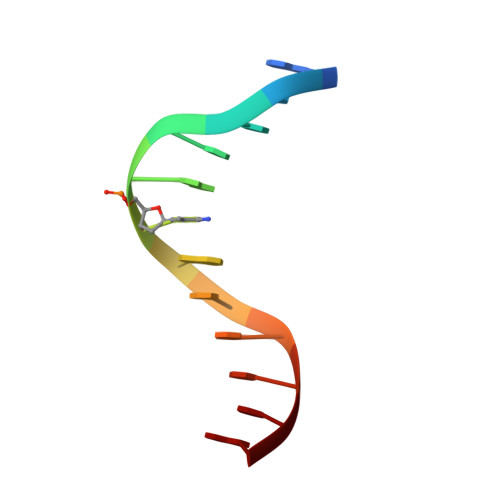
Structural basis for the ability of MBD domains to bind methyl-CG and TG sites in DNA.
Liu, K., Xu, C., Lei, M., Yang, A., Loppnau, P., Hughes, T.R., Min, J.(2018) J Biological Chem 293: 7344-7354
- PubMed: 29567833
- DOI: https://doi.org/10.1074/jbc.RA118.001785
- Primary Citation of Related Structures:
6C1A, 6C1T, 6C1U, 6C1V, 6CNP, 6CNQ - PubMed Abstract:
Cytosine methylation is a well-characterized epigenetic mark and occurs at both CG and non-CG sites in DNA. Both methylated CG (mCG)- and mCH (H = A, C, or T)-containing DNAs, especially mCAC-containing DNAs, are recognized by methyl-CpG-binding protein 2 (MeCP2) to regulate gene expression in neuron development. However, the molecular mechanism involved in the binding of methyl-CpG-binding domain (MBD) of MeCP2 to these different DNA motifs is unclear. Here, we systematically characterized the DNA-binding selectivities of the MBD domains in MeCP2 and MBD1-4 with isothermal titration calorimetry-based binding assays, mutagenesis studies, and X-ray crystallography. We found that the MBD domains of MeCP2 and MBD1-4 bind mCG-containing DNAs independently of the sequence identity outside the mCG dinucleotide. Moreover, some MBD domains bound to both methylated and unmethylated CA dinucleotide-containing DNAs, with a preference for the CAC sequence motif. We also found that the MBD domains bind to mCA or nonmethylated CA DNA by recognizing the complementary TG dinucleotide, which is consistent with an overlooked ligand of MeCP2, i.e. the matrix/scaffold attachment regions (MARs/SARs) with a consensus sequence of 5'-GGTGT-3' that was identified in early 1990s. Our results also explain why MeCP2 exhibits similar binding affinity to both mCA- and hmCA-containing dsDNAs. In summary, our results suggest that in addition to mCG sites, unmethylated CA or TG sites also serve as DNA-binding sites for MeCP2 and other MBD-containing proteins. This discovery expands the genome-wide activity of MBD-containing proteins in gene regulation.
- Structural Genomics Consortium, University of Toronto, Toronto, Ontario M5G 1L7; Department of Physiology, University of Toronto, Toronto, Ontario M5S 1A8, Canada.
Organizational Affiliation: