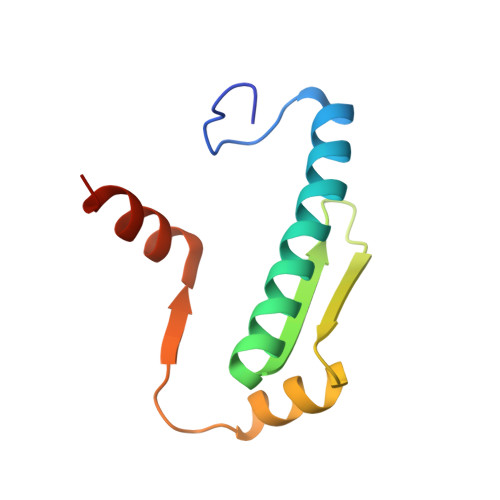
The Cancer Mutation D83V Induces an alpha-Helix to beta-Strand Conformation Switch in MEF2B.
Lei, X., Kou, Y., Fu, Y., Rajashekar, N., Shi, H., Wu, F., Xu, J., Luo, Y., Chen, L.(2018) J Mol Biology 430: 1157-1172
- PubMed: 29477338
- DOI: https://doi.org/10.1016/j.jmb.2018.02.012
- Primary Citation of Related Structures:
6BYY, 6BZ1 - PubMed Abstract:
MEF2B is a major target of somatic mutations in non-Hodgkin lymphoma. Most of these mutations are non-synonymous substitutions of surface residues in the MADS-box/MEF2 domain. Among them, D83V is the most frequent mutation found in tumor cells. The link between this hotspot mutation and cancer is not well understood. Here we show that the D83V mutation induces a dramatic α-helix to β-strand switch in the MEF2 domain. Located in an α-helix region rich in β-branched residues, the D83V mutation not only removes the extensive helix stabilization interactions but also introduces an additional β-branched residue that further shifts the conformation equilibrium from α-helix to β-strand. Cross-database analyses of cancer mutations and chameleon sequences revealed a number of well-known cancer targets harboring β-strand favoring mutations in chameleon α-helices, suggesting a commonality of such conformational switch in certain cancers and a new factor to consider when stratifying the rapidly expanding cancer mutation data.
- Molecular and Computational Biology, Department of Biological Sciences, University of Southern California, Los Angeles, CA 90089, USA.
Organizational Affiliation: