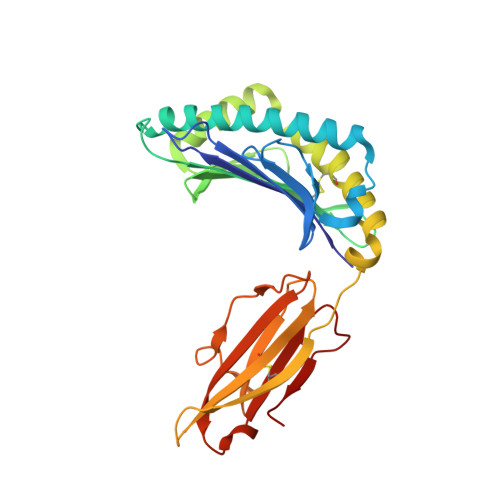
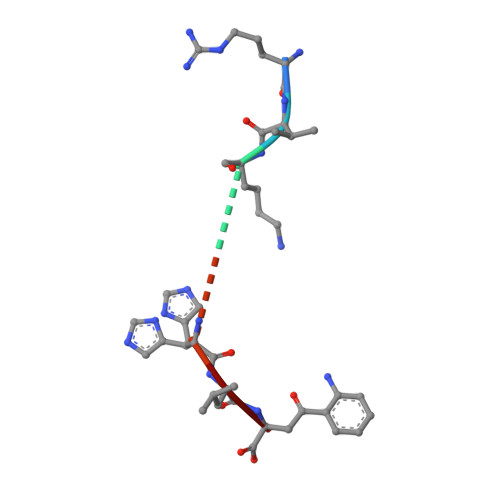
Identification of Native and Posttranslationally Modified HLA-B*57:01-Restricted HIV Envelope Derived Epitopes Using Immunoproteomics.
Ramarathinam, S.H., Gras, S., Alcantara, S., Yeung, A.W.S., Mifsud, N.A., Sonza, S., Illing, P.T., Glaros, E.N., Center, R.J., Thomas, S.R., Kent, S.J., Ternette, N., Purcell, D.F.J., Rossjohn, J., Purcell, A.W.(2018) Proteomics 18: e1700253-e1700253
- PubMed: 29437277
- DOI: https://doi.org/10.1002/pmic.201700253
- Primary Citation of Related Structures:
6BXP, 6BXQ - PubMed Abstract:
The recognition of pathogen-derived peptides by T lymphocytes is the cornerstone of adaptive immunity, whereby intracellular antigens are degraded in the cytosol and short peptides assemble with class I human leukocyte antigen (HLA) molecules in the ER. These peptide-HLA complexes egress to the cell surface and are scrutinized by cytotoxic CD8+ T-cells leading to the eradication of the infected cell. Here, naturally presented HLA-B*57:01 bound peptides derived from the envelope protein of the human immunodeficiency virus (HIVenv) are identified. HIVenv peptides are present at a very small percentage of the overall HLA-B*57:01 peptidome (<0.1%) and both native and posttranslationally modified forms of two distinct HIV peptides are identified. Notably, a peptide bearing a natively encoded C-terminal tryptophan residue is also present in a modified form containing a kynurenine residue. Kynurenine is a major product of tryptophan catabolism and is abundant during inflammation and infection. Binding of these peptides at a molecular level and their immunogenicity in preliminary functional studies are examined. Modest immune responses are observed to the modified HIVenv peptide, highlighting a potential role for kynurenine-modified peptides in the immune response to HIV and other viral infections.
- Infection and Immunity Program, Biomedicine Discovery Institute & Department of Biochemistry and Molecular Biology, Monash University, Clayton, Australia.
Organizational Affiliation: