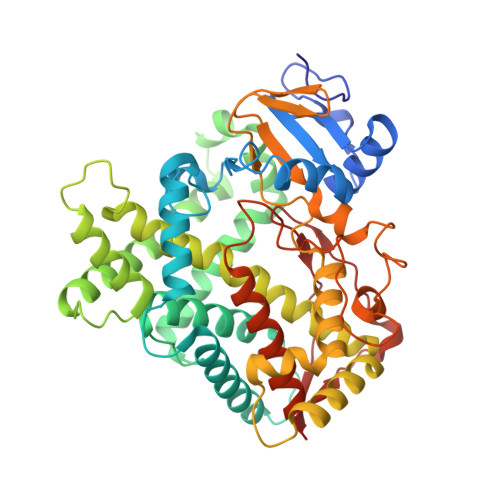
Structure of cytochrome P450 2B4 with an acetate ligand and an active site hydrogen bond network similar to oxyferrous P450cam.
Yang, Y., Bu, W., Im, S., Meagher, J., Stuckey, J., Waskell, L.(2018) J Inorg Biochem 185: 17-25
- PubMed: 29730233
- DOI: https://doi.org/10.1016/j.jinorgbio.2018.04.015
- Primary Citation of Related Structures:
6BWW - Department of Anesthesiology, University of Michigan and VA Medical Research Center, 2215 Fuller Rd, Ann Arbor, MI 48105, USA.
Organizational Affiliation: