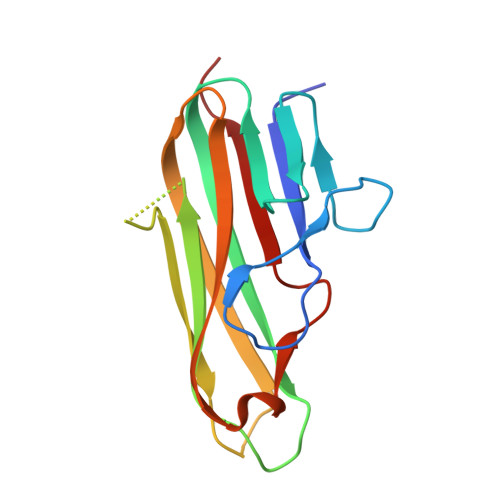
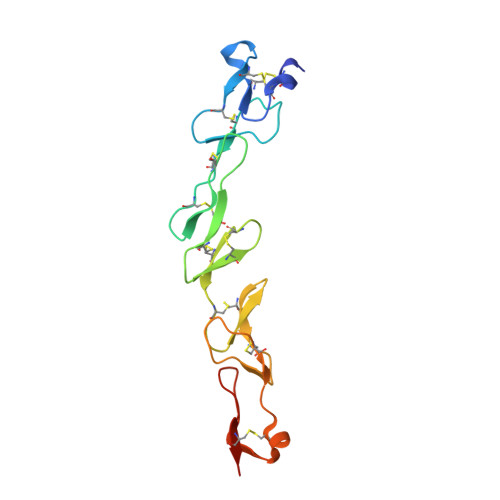
Crystal structure of the human 4-1BB/4-1BBL complex.
Gilbreth, R.N., Oganesyan, V.Y., Amdouni, H., Novarra, S., Grinberg, L., Barnes, A., Baca, M.(2018) J Biological Chem 293: 9880-9891
- PubMed: 29720399
- DOI: https://doi.org/10.1074/jbc.RA118.002803
- Primary Citation of Related Structures:
6BWV - PubMed Abstract:
4-1BBL is a member of the tumor necrosis factor (TNF) superfamily and is the ligand for the TNFR superfamily receptor, 4-1BB. 4-1BB plays an immunomodulatory role in T cells and NK cells, and agonists of this receptor have garnered strong attention as potential immunotherapy agents. Broadly speaking, the structural features of TNF superfamily members, their receptors, and ligand-receptor complexes are similar. However, a published crystal structure of human 4-1BBL suggests that it may be unique in this regard, exhibiting a three-bladed propeller-like trimer assembly that is distinctly different from that observed in other family members. This unusual structure also suggests that the human 4-1BB/4-1BBL complex may be structurally unique within the TNF/TNFR superfamily, but to date no structural data have been reported. Here we report the crystal structure of the human 4-1BB/4-1BBL complex at 2.4-Å resolution. In this structure, 4-1BBL does not adopt the unusual trimer assembly previously reported, but instead forms a canonical bell-shaped trimer typical of other TNF superfamily members. The structure of 4-1BB is also largely canonical as is the 4-1BB/4-1BBL complex. Mutational data support the 4-1BBL structure reported here as being biologically relevant, suggesting that the previously reported structure is not. Together, the data presented here offer insight into structure/function relationships in the 4-1BB/4-1BBL system and improve our structural understanding of the TNF/TNFR superfamily more broadly.
- From the Department of Antibody Discovery and Protein Engineering, MedImmune LLC, Gaithersburg, Maryland 20878 gilbrethr@medimmune.com.
Organizational Affiliation: