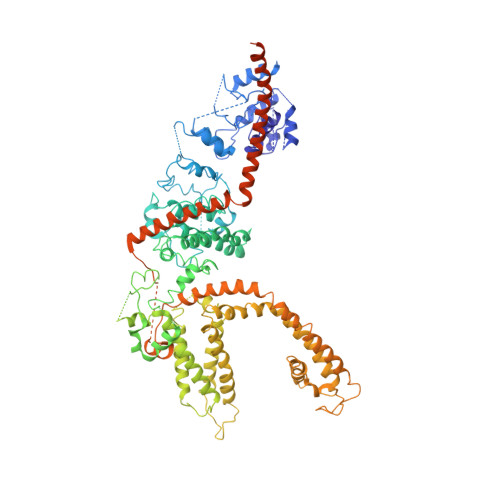
Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Duan, J., Li, Z., Li, J., Hulse, R.E., Santa-Cruz, A., Valinsky, W.C., Abiria, S.A., Krapivinsky, G., Zhang, J., Clapham, D.E.(2018) Proc Natl Acad Sci U S A 115: E8201-E8210
- PubMed: 30108148
- DOI: https://doi.org/10.1073/pnas.1810719115
- Primary Citation of Related Structures:
5ZX5, 6BWD, 6BWF - PubMed Abstract:
The transient receptor potential ion channel subfamily M, member 7 (TRPM7), is a ubiquitously expressed protein that is required for mouse embryonic development. TRPM7 contains both an ion channel and an α-kinase. The channel domain comprises a nonselective cation channel with notable permeability to Mg 2+ and Zn 2+ Here, we report the closed state structures of the mouse TRPM7 channel domain in three different ionic conditions to overall resolutions of 3.3, 3.7, and 4.1 Å. The structures reveal key residues for an ion binding site in the selectivity filter, with proposed partially hydrated Mg 2+ ions occupying the center of the conduction pore. In high [Mg 2+ ], a prominent external disulfide bond is found in the pore helix, which is essential for ion channel function. Our results provide a structural framework for understanding the TRPM1/3/6/7 subfamily and extend the knowledge base upon which to study the diversity and evolution of TRP channels.
- Department of Cardiology, Boston Children's Hospital, Boston, MA 02115.
Organizational Affiliation: