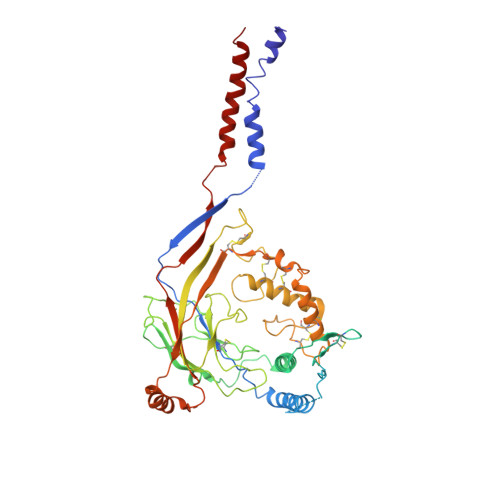
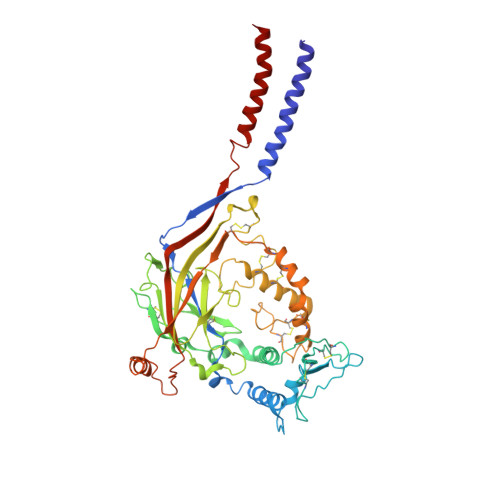
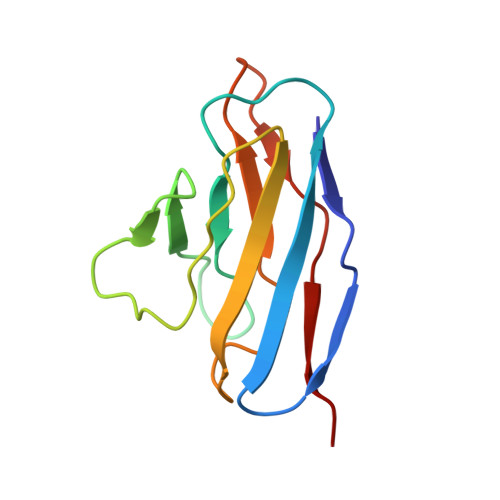
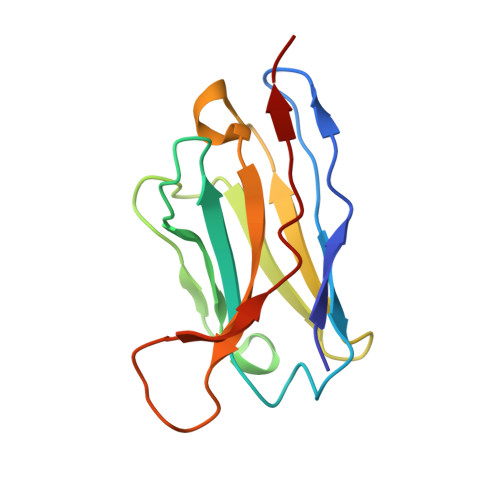
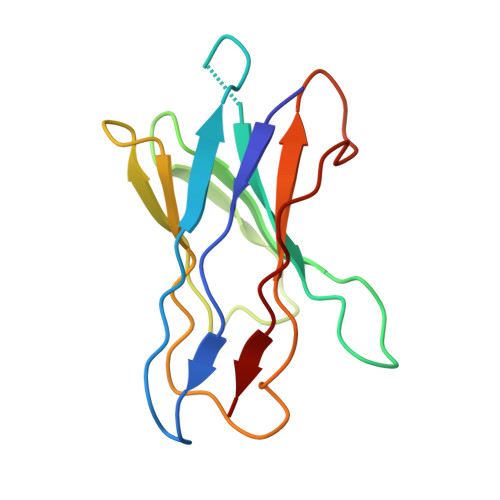
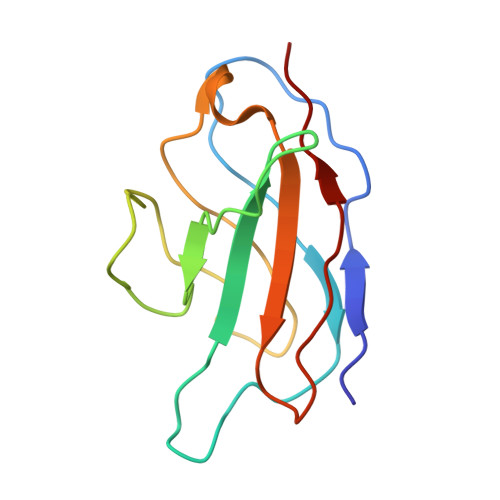
Structure of the human epithelial sodium channel by cryo-electron microscopy.
Noreng, S., Bharadwaj, A., Posert, R., Yoshioka, C., Baconguis, I.(2018) Elife 7
- PubMed: 30251954
- DOI: https://doi.org/10.7554/eLife.39340
- Primary Citation of Related Structures:
6BQN - PubMed Abstract:
The epithelial sodium channel (ENaC), a member of the ENaC/DEG superfamily, regulates Na + and water homeostasis. ENaCs assemble as heterotrimeric channels that harbor protease-sensitive domains critical for gating the channel. Here, we present the structure of human ENaC in the uncleaved state determined by single-particle cryo-electron microscopy. The ion channel is composed of a large extracellular domain and a narrow transmembrane domain. The structure reveals that ENaC assembles with a 1:1:1 stoichiometry of α:β:γ subunits arranged in a counter-clockwise manner. The shape of each subunit is reminiscent of a hand with key gating domains of a 'finger' and a 'thumb.' Wedged between these domains is the elusive protease-sensitive inhibitory domain poised to regulate conformational changes of the 'finger' and 'thumb'; thus, the structure provides the first view of the architecture of inhibition of ENaC.
- Department of Biochemistry & Molecular Biology, Oregon Health and Science University, Portland, United States.
Organizational Affiliation: