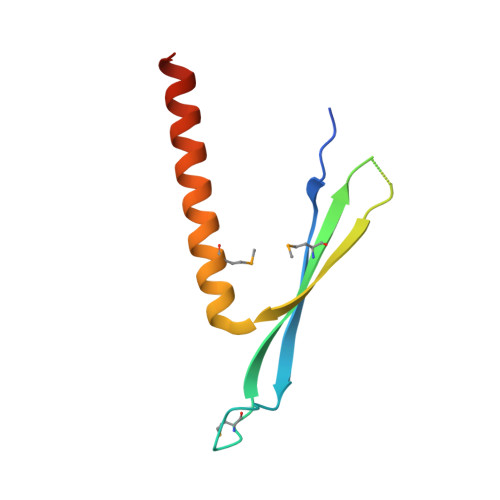
Structural insights into the architecture and membrane interactions of the conserved COMMD proteins.
Healy, M.D., Hospenthal, M.K., Hall, R.J., Chandra, M., Chilton, M., Tillu, V., Chen, K.E., Celligoi, D.J., McDonald, F.J., Cullen, P.J., Lott, J.S., Collins, B.M., Ghai, R.(2018) Elife 7
- PubMed: 30067224
- DOI: https://doi.org/10.7554/eLife.35898
- Primary Citation of Related Structures:
4NKN, 4OE9, 6BP6 - PubMed Abstract:
The COMMD proteins are a conserved family of proteins with central roles in intracellular membrane trafficking and transcription. They form oligomeric complexes with each other and act as components of a larger assembly called the CCC complex, which is localized to endosomal compartments and mediates the transport of several transmembrane cargos. How these complexes are formed however is completely unknown. Here, we have systematically characterised the interactions between human COMMD proteins, and determined structures of COMMD proteins using X-ray crystallography and X-ray scattering to provide insights into the underlying mechanisms of homo- and heteromeric assembly. All COMMD proteins possess an α-helical N-terminal domain, and a highly conserved C-terminal domain that forms a tightly interlocked dimeric structure responsible for COMMD-COMMD interactions. The COMM domains also bind directly to components of CCC and mediate non-specific membrane association. Overall these studies show that COMMD proteins function as obligatory dimers with conserved domain architectures.
- Institute for Molecular Bioscience, The University of Queensland, St. Lucia, Australia.
Organizational Affiliation: