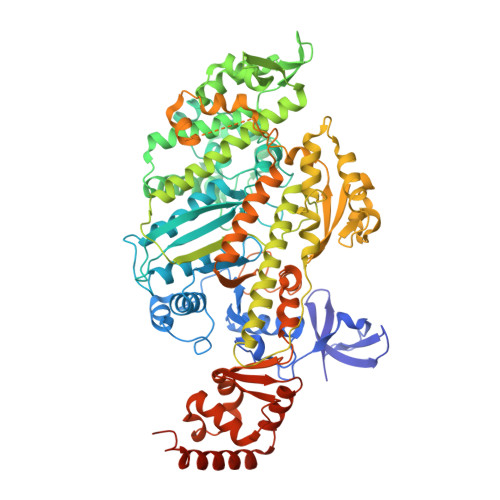
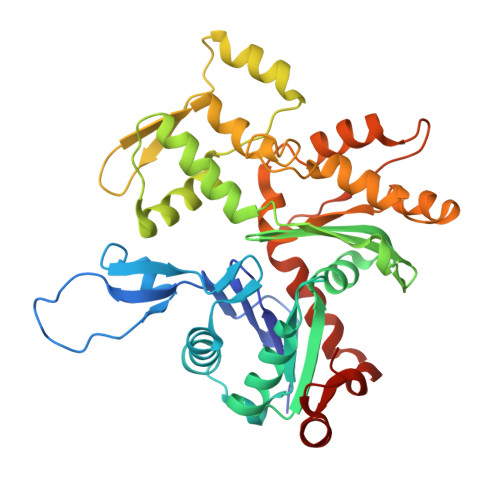
Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity.
Gurel, P.S., Kim, L.Y., Ruijgrok, P.V., Omabegho, T., Bryant, Z., Alushin, G.M.(2017) Elife 6
- PubMed: 29199952
- DOI: https://doi.org/10.7554/eLife.31125
- Primary Citation of Related Structures:
6BNO, 6BNP, 6BNQ, 6BNU, 6BNV, 6BNW - PubMed Abstract:
Despite extensive scrutiny of the myosin superfamily, the lack of high-resolution structures of actin-bound states has prevented a complete description of its mechanochemical cycle and limited insight into how sequence and structural diversification of the motor domain gives rise to specialized functional properties. Here we present cryo-EM structures of the unique minus-end directed myosin VI motor domain in rigor (4.6 Å) and Mg-ADP (5.5 Å) states bound to F-actin. Comparison to the myosin IIC-F-actin rigor complex reveals an almost complete lack of conservation of residues at the actin-myosin interface despite preservation of the primary sequence regions composing it, suggesting an evolutionary path for motor specialization. Additionally, analysis of the transition from ADP to rigor provides a structural rationale for force sensitivity in this step of the mechanochemical cycle. Finally, we observe reciprocal rearrangements in actin and myosin accompanying the transition between these states, supporting a role for actin structural plasticity during force generation by myosin VI.
- Laboratory of Structural Biophysics and Mechanobiology, The Rockefeller University, New York, United States.
Organizational Affiliation: