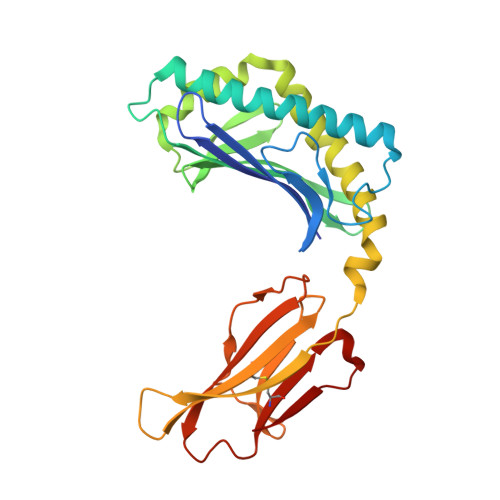
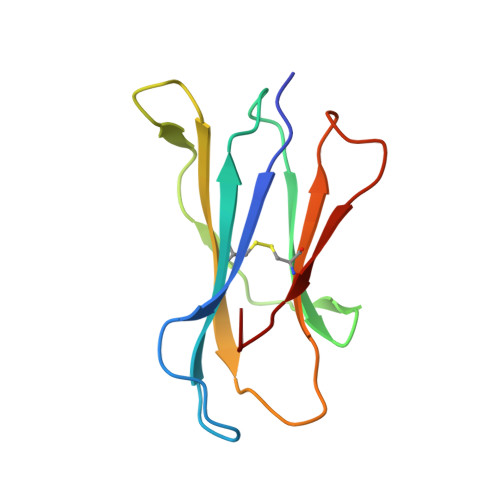
Differing roles of CD1d2 and CD1d1 proteins in type I natural killer T cell development and function.
Sundararaj, S., Zhang, J., Krovi, S.H., Bedel, R., Tuttle, K.D., Veerapen, N., Besra, G.S., Khandokar, Y., Praveena, T., Le Nours, J., Matsuda, J.L., Rossjohn, J., Gapin, L.(2018) Proc Natl Acad Sci U S A 115: E1204-E1213
- PubMed: 29351991
- DOI: https://doi.org/10.1073/pnas.1716669115
- Primary Citation of Related Structures:
6BMH, 6BMK - PubMed Abstract:
MHC class I-like CD1 molecules have evolved to present lipid-based antigens to T cells. Differences in the antigen-binding clefts of the CD1 family members determine the conformation and size of the lipids that are presented, although the factors that shape CD1 diversity remain unclear. In mice, two homologous genes, CD1D1 and CD1D2, encode the CD1d protein, which is essential to the development and function of natural killer T (NKT) cells. However, it remains unclear whether both CD1d isoforms are equivalent in their antigen presentation capacity and functions. Here, we report that CD1d2 molecules are expressed in the thymus of some mouse strains, where they select functional type I NKT cells. Intriguingly, the T cell antigen receptor repertoire and phenotype of CD1d2-selected type I NKT cells in CD1D1 -/- mice differed from CD1d1-selected type I NKT cells. The structures of CD1d2 in complex with endogenous lipids and a truncated acyl-chain analog of α-galactosylceramide revealed that its A'-pocket was restricted in size compared with CD1d1. Accordingly, CD1d2 molecules could not present glycolipid antigens with long acyl chains efficiently, favoring the presentation of short acyl chain antigens. These results indicate that the two CD1d molecules present different sets of self-antigen(s) in the mouse thymus, thereby impacting the development of invariant NKT cells.
- Infection and Immunity Program, Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia.
Organizational Affiliation: