Stable human IgG antibody therapeutics with native framework structure
Christie, M., Rouet, R., Nevoltris, D., Langley, D., Schofield, P., Fabri, L., Tingey, K., Lowe, D., Jermutus, L., Christ, D.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Avastin Light Chain Fab fragment mutant | A [auth B], C [auth L] | 214 | Homo sapiens | Mutation(s): 0 | 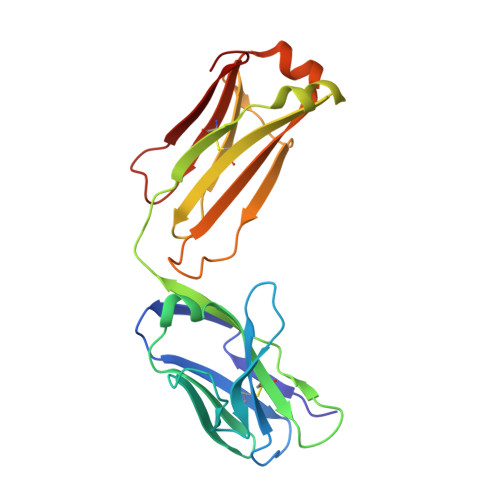 |
UniProt | |||||
Find proteins for Q8TCD0 (Homo sapiens) Explore Q8TCD0 Go to UniProtKB: Q8TCD0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8TCD0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Avastin Heavy Chain Fab fragment mutant | B [auth A], D [auth H] | 231 | Homo sapiens | Mutation(s): 0 | 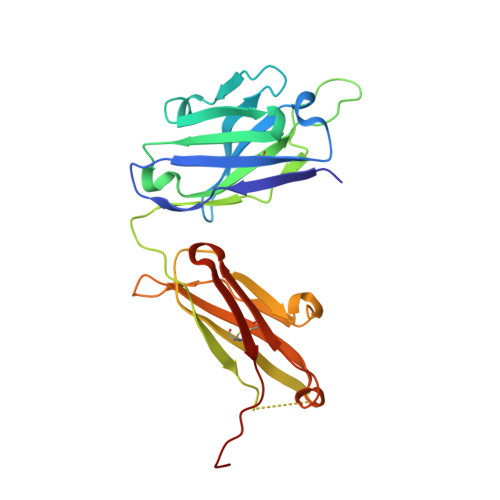 |
UniProt | |||||
Find proteins for P0DOX5 (Homo sapiens) Explore P0DOX5 Go to UniProtKB: P0DOX5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DOX5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Vascular endothelial growth factor A | E [auth G], F [auth C] | 102 | Homo sapiens | Mutation(s): 0 Gene Names: VEGFA, VEGF | 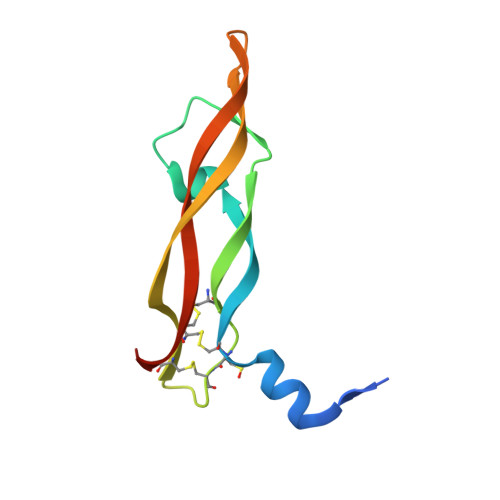 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P15692 (Homo sapiens) Explore P15692 Go to UniProtKB: P15692 | |||||
PHAROS: P15692 GTEx: ENSG00000112715 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P15692 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MES Query on MES | I [auth H], J [auth H] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | G [auth B], H [auth L] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSO Query on CSO | E [auth G], F [auth C] | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 83.279 | α = 90 |
| b = 88.749 | β = 111.87 |
| c = 106.676 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Australian Research Council (ARC) | Australia | DE160100608 |
| Australian Research Council (ARC) | Australia | DP160104915 |