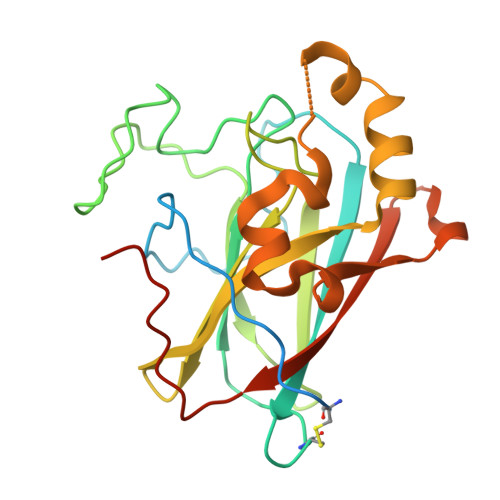
Structural and functional characterization of an intradiol ring-cleavage dioxygenase from the polyphagous spider mite herbivore Tetranychus urticae Koch.
Schlachter, C.R., Daneshian, L., Amaya, J., Klapper, V., Wybouw, N., Borowski, T., Van Leeuwen, T., Grbic, V., Grbic, M., Makris, T.M., Chruszcz, M.(2019) Insect Biochem Mol Biol 107: 19-30
- PubMed: 30529144
- DOI: https://doi.org/10.1016/j.ibmb.2018.12.001
- Primary Citation of Related Structures:
5VG2, 6BDJ - PubMed Abstract:
Genome analyses of the polyphagous spider mite herbivore Tetranychus urticae (two-spotted spider mite) revealed the presence of a set of 17 genes that code for secreted proteins belonging to the "intradiol dioxygenase-like" subgroup. Phylogenetic analyses indicate that this novel enzyme family has been acquired by horizontal gene transfer. In order to better understand the role of these proteins in T. urticae, we have structurally and functionally characterized one paralog (tetur07g02040). It was demonstrated that this protein is indeed an intradiol ring-cleavage dioxygenase, as the enzyme is able to cleave catechol between two hydroxyl-groups using atmospheric dioxygen. The enzyme was characterized functionally and structurally. The active site of the T. urticae enzyme contains an Fe 3+ cofactor that is coordinated by two histidine and two tyrosine residues, an arrangement that is similar to those observed in bacterial homologs. However, the active site is significantly more solvent exposed than in bacterial proteins. Moreover, the mite enzyme is monomeric, while almost all structurally characterized bacterial homologs form oligomeric assemblies. Tetur07g02040 is not only the first spider mite dioxygenase that has been characterized at the molecular level, but is also the first structurally characterized intradiol ring-cleavage dioxygenase originating from a eukaryote.
- Department of Chemistry and Biochemistry, University of South Carolina, Columbia, SC, 29208, USA.
Organizational Affiliation: