I-LtrI E29D bound to cognate substrate (nicked complex)
McMurrough, T.A., Brown, C., Zhang, K., Junop, M., Gloor, G.B., Edgell, D.R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosomal protein 3/homing endonuclease-like fusion protein | A, E [auth D], I [auth H], M [auth L] | 315 | Leptographium truncatum | Mutation(s): 1 Gene Names: HEG fusion, rps3 | 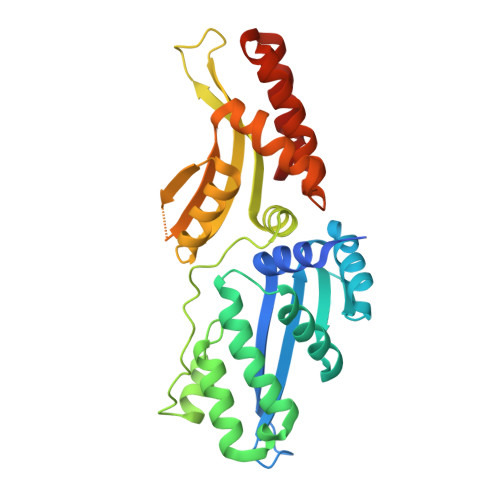 |
UniProt | |||||
Find proteins for C7SWF3 (Leptographium truncatum) Explore C7SWF3 Go to UniProtKB: C7SWF3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C7SWF3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*GP*GP*TP*CP*TP*AP*AP*AP*CP*GP*TP*CP*GP*TP*AP*T)-3') | B, F [auth E], J [auth I], N | 16 | Leptographium truncatum | 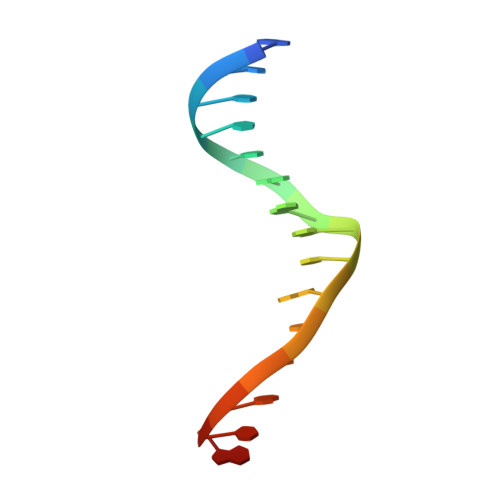 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (26-MER) | C, G [auth F], K [auth J], O | 26 | Leptographium truncatum |  | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*TP*AP*GP*GP*AP*GP*CP*AP*TP*TP*T)-3') | D [auth X], H [auth G], L [auth K], P | 11 | Leptographium truncatum |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 43.76 | α = 90.03 |
| b = 66.315 | β = 90.03 |
| c = 169.924 | γ = 90.22 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| MOSFLM | data reduction |
| Aimless | data scaling |
| PHASER | phasing |