Synthesis and Pharmacological Characterization of C4beta-Amide-Substituted 2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1 S,2 S,4 S,5 R,6 S)-2-Amino-4-[(3-methoxybenzoyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2794193), a Highly Potent and Selective mGlu3Receptor Agonist.
Monn, J.A., Henry, S.S., Massey, S.M., Clawson, D.K., Chen, Q., Diseroad, B.A., Bhardwaj, R.M., Atwell, S., Lu, F., Wang, J., Russell, M., Heinz, B.A., Wang, X.S., Carter, J.H., Getman, B.G., Adragni, K., Broad, L.M., Sanger, H.E., Ursu, D., Catlow, J.T., Swanson, S., Johnson, B.G., Shaw, D.B., McKinzie, D.L., Hao, J.(2018) J Med Chem 61: 2303-2328
- PubMed: 29350927
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01481
- Primary Citation of Related Structures:
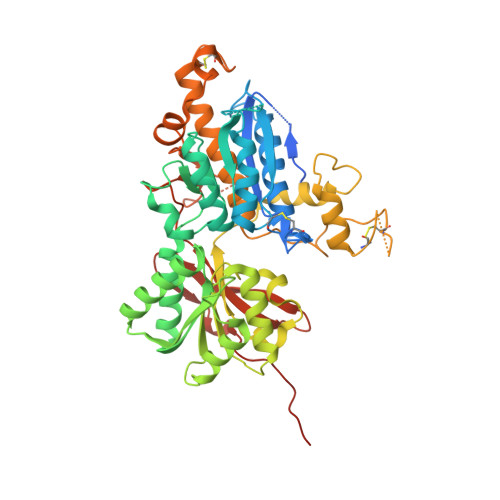
6B7H - PubMed Abstract:
Multiple therapeutic opportunities have been suggested for compounds capable of selective activation of metabotropic glutamate 3 (mGlu 3 ) receptors, but small molecule tools are lacking. As part of our ongoing efforts to identify potent, selective, and systemically bioavailable agonists for mGlu 2 and mGlu 3 receptor subtypes, a series of C4 β -N-linked variants of (1 S,2 S,5 R,6 S)-2-amino-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid 1 (LY354740) were prepared and evaluated for both mGlu 2 and mGlu 3 receptor binding affinity and functional cellular responses. From this investigation we identified (1 S,2 S,4 S,5 R,6 S)-2-amino-4-[(3-methoxybenzoyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid 8p (LY2794193), a molecule that demonstrates remarkable mGlu 3 receptor selectivity. Crystallization of 8p with the amino terminal domain of hmGlu 3 revealed critical binding interactions for this ligand with residues adjacent to the glutamate binding site, while pharmacokinetic assessment of 8p combined with its effect in an mGlu 2 receptor-dependent behavioral model provides estimates for doses of this compound that would be expected to selectively engage and activate central mGlu 3 receptors in vivo.