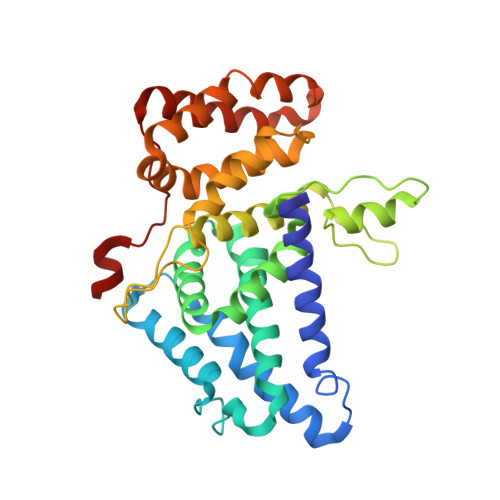
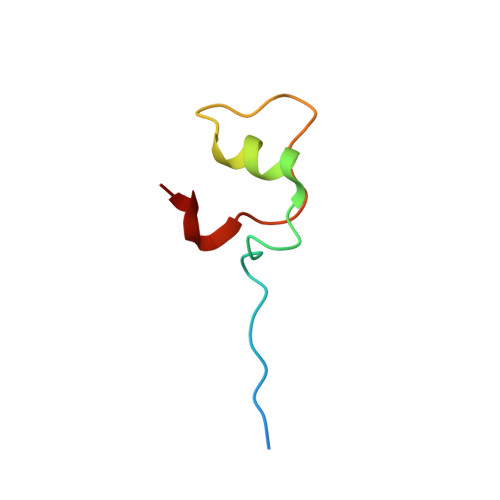
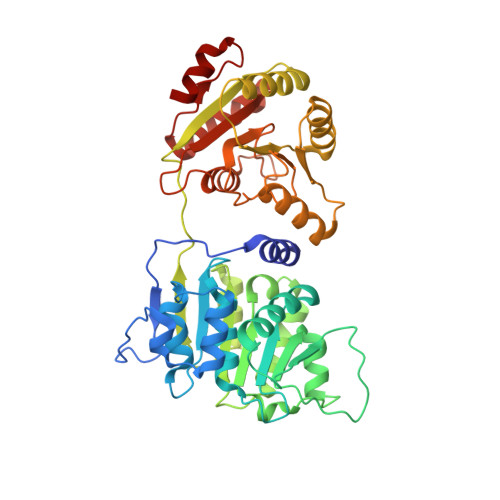
Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Lin, D.H., Correia, A.R., Cai, S.W., Huber, F.M., Jette, C.A., Hoelz, A.(2018) Nat Commun 9: 2319-2319
- PubMed: 29899397
- DOI: https://doi.org/10.1038/s41467-018-04459-3
- Primary Citation of Related Structures:
6B4E, 6B4F, 6B4G, 6B4H, 6B4I, 6B4J, 6B4K - PubMed Abstract:
The nuclear pore complex (NPC) controls the passage of macromolecules between the nucleus and cytoplasm, but how the NPC directly participates in macromolecular transport remains poorly understood. In the final step of mRNA export, the DEAD-box helicase DDX19 is activated by the nucleoporins Gle1, Nup214, and Nup42 to remove Nxf1•Nxt1 from mRNAs. Here, we report crystal structures of Gle1•Nup42 from three organisms that reveal an evolutionarily conserved binding mode. Biochemical reconstitution of the DDX19 ATPase cycle establishes that human DDX19 activation does not require IP 6 , unlike its fungal homologs, and that Gle1 stability affects DDX19 activation. Mutations linked to motor neuron diseases cause decreased Gle1 thermostability, implicating nucleoporin misfolding as a disease determinant. Crystal structures of human Gle1•Nup42•DDX19 reveal the structural rearrangements in DDX19 from an auto-inhibited to an RNA-binding competent state. Together, our results provide the foundation for further mechanistic analyses of mRNA export in humans.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, 1200 East California Boulevard, Pasadena, CA, 91125, USA.
Organizational Affiliation: