Bacterially derived synthetic mimetics of mammalian oligomannose prime antibody responses that neutralize HIV infectivity.
Pantophlet, R., Trattnig, N., Murrell, S., Lu, N., Chau, D., Rempel, C., Wilson, I.A., Kosma, P.(2017) Nat Commun 8: 1601-1601
- PubMed: 29150603
- DOI: https://doi.org/10.1038/s41467-017-01640-y
- Primary Citation of Related Structures:
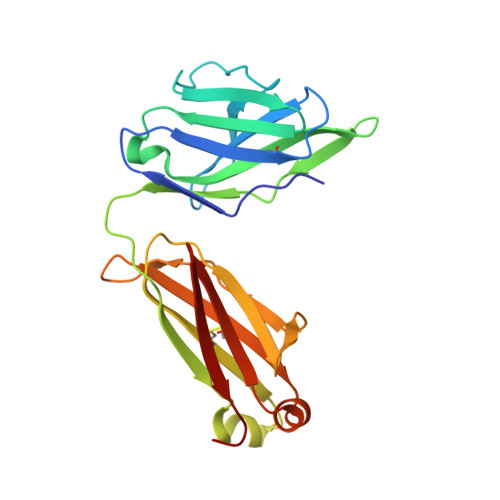
6B3D - PubMed Abstract:
Oligomannose-type glycans are among the major targets on the gp120 component of the HIV envelope protein (Env) for broadly neutralizing antibodies (bnAbs). However, attempts to elicit oligomannose-specific nAbs by immunizing with natural or synthetic oligomannose have so far not been successful, possibly due to B cell tolerance checkpoints. Here we design and synthesize oligomannose mimetics, based on the unique chemical structure of a recently identified bacterial lipooligosaccharide, to appear foreign to the immune system. One of these mimetics is bound avidly by members of a family of oligomannose-specific bnAbs and their putative common germline precursor when presented as a glycoconjugate. The crystal structure of one of the mimetics bound to a member of this bnAb family confirms the antigenic resemblance. Lastly, immunization of human-antibody transgenic animals with a lead mimetic evokes nAbs with specificities approaching those of existing bnAbs. These results provide evidence for utilizing antigenic mimicry to elicit oligomannose-specific bnAbs to HIV-1.
- Faculty of Health Sciences, Simon Fraser University, Burnaby, BC, Canada, V5A1S6. rpantophlet@sfu.ca.
Organizational Affiliation: