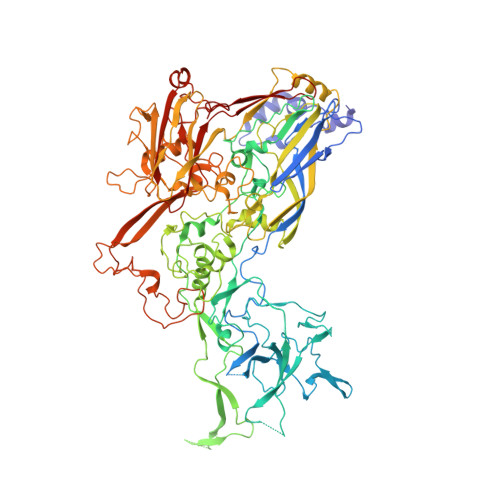
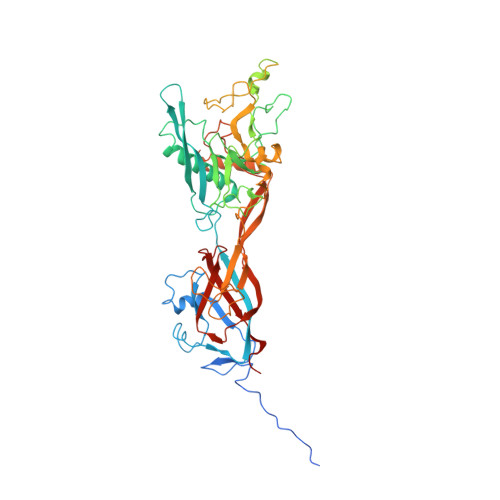
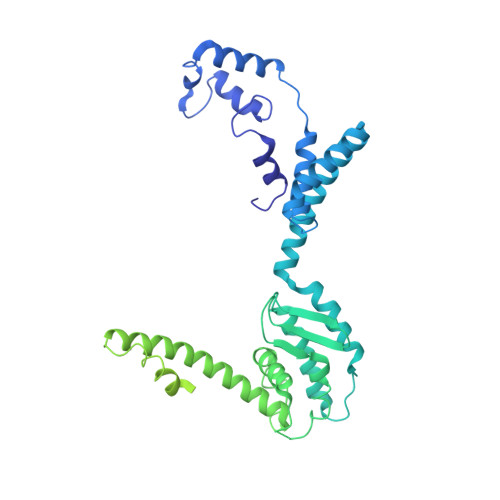
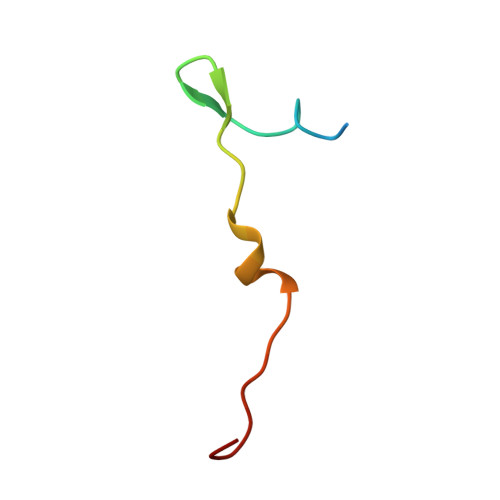
Atomic Structures of Minor Proteins VI and VII in Human Adenovirus.
Dai, X., Wu, L., Sun, R., Zhou, Z.H.(2017) J Virol 91
- PubMed: 28978703
- DOI: https://doi.org/10.1128/JVI.00850-17
- Primary Citation of Related Structures:
6B1T - PubMed Abstract:
Human adenoviruses (Ad) are double-stranded DNA (dsDNA) viruses associated with infectious diseases, but they are better known as tools for gene delivery and oncolytic anticancer therapy. Atomic structures of Ad provide the basis for the development of antivirals and for engineering efforts toward more effective applications. Since 2010, atomic models of human Ad5 have been derived independently from photographic film cryo-electron microscopy (cryo-EM) and X-ray crystallography studies, but discrepancies exist concerning the assignment of cement proteins IIIa, VIII, and IX. To clarify these discrepancies, we employed the technology of direct electron counting to obtain a cryo-EM structure of human Ad5 at 3.2-Å resolution. Our improved structure unambiguously confirms our previous cryo-EM models of proteins IIIa, VIII, and IX and explains the likely cause of conflict in the crystallography models. The improved structure also allows the identification of three new components in the cavity of hexon-the cleaved N terminus of precursor protein VI (pVIn), the cleaved N terminus of precursor protein VII (pVIIn2), and mature protein VI. The binding of pVIIn2-and, by extension, that of genome-condensing pVII-to hexons is consistent with the previously proposed dsDNA genome-capsid coassembly for adenoviruses, which resembles that of single-stranded RNA (ssRNA) viruses but differs from the well-established mechanism of pumping dsDNA into a preformed protein capsid exemplified by tailed bacteriophages and herpesviruses. IMPORTANCE Adenovirus is a double-edged sword to humans: it is a widespread pathogen but can be used as a bioengineering tool for anticancer and gene therapies. The atomic structure of the virus provides the basis for antiviral and application developments, but conflicting atomic models for the important cement proteins IIIa, VIII, and IX from conventional/film cryo-EM and X-ray crystallography studies have caused confusion. Using cutting-edge cryo-EM technology with electron counting, we improved the structure of human adenovirus type 5 and confirmed our previous models of cement proteins IIIa, VIII, and IX, thus clarifying the inconsistent structures. The improved structure also reveals atomic details of membrane-lytic protein VI and genome-condensing protein VII and supports the previously proposed genome-capsid coassembly mechanism for adenoviruses.
- Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles, Los Angeles, California, USA.
Organizational Affiliation: