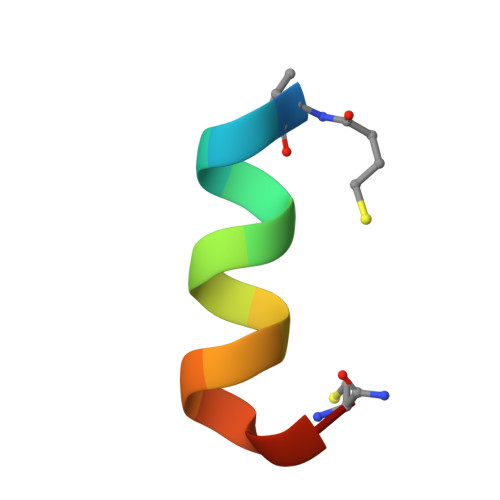
Design of a Short Thermally Stable alpha-Helix Embedded in a Macrocycle.
Wu, H., Acharyya, A., Wu, Y., Liu, L., Jo, H., Gai, F., DeGrado, W.F.(2018) Chembiochem 19: 902-906
- PubMed: 29417711
- DOI: https://doi.org/10.1002/cbic.201800026
- Primary Citation of Related Structures:
6ANF, 6B17 - PubMed Abstract:
Although helices play key roles in peptide-protein and protein-protein interactions, the helical conformation is generally unstable for short peptides (10-15 residues) in aqueous solution in the absence of their binding partners. Thus, stabilizing the helical conformation of peptides can lead to increases in binding potency, specificity, and stability towards proteolytic degradation. Helices have been successfully stabilized by introducing side chain-to-side chain crosslinks within the central portion of the helix. However, this approach leaves the ends of the helix free, thus leading to fraying and exposure of the non-hydrogen-bonded amide groups to solvent. Here, we develop a "capped-strapped" peptide strategy to stabilize helices by embedding the entire length of the helix within a macrocycle, which also includes a semirigid organic template as well as end-capping interactions. We have designed a ten-residue capped-strapped helical peptide that behaves like a miniprotein, with a cooperative thermal unfolding transition and T m ≈70 °C, unprecedented for helical peptides of this length. The NMR structure determination confirmed the design, and X-ray crystallography revealed a novel quaternary structure with implications for foldamer design.
- Department of Pharmaceutical Chemistry, University of California, San Francisco, CA, 94158, USA.
Organizational Affiliation: