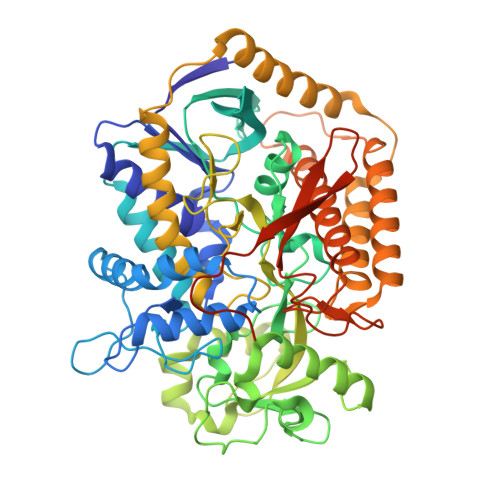
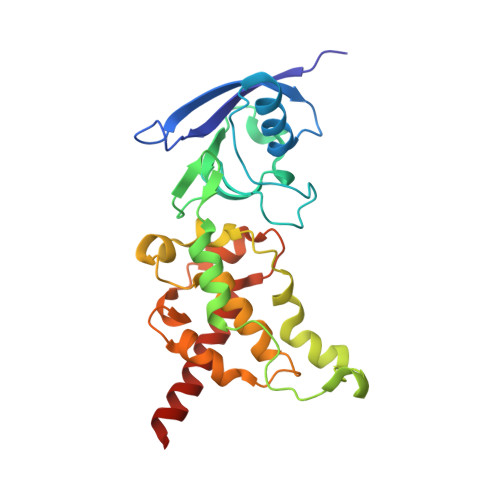
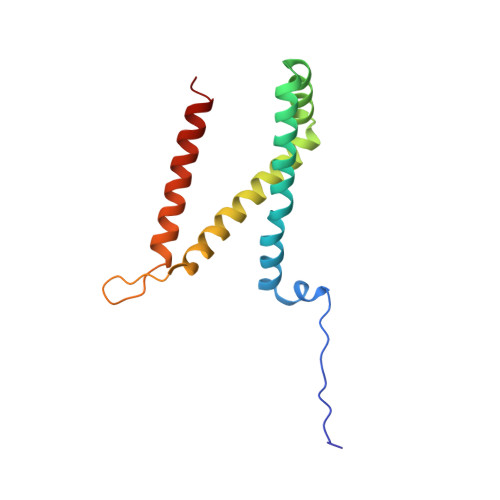
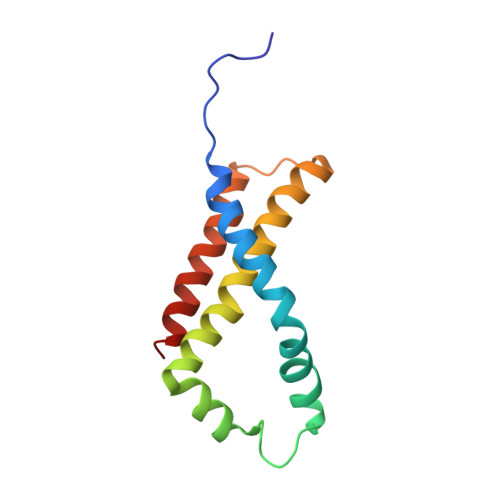
New crystal forms of the integral membrane Escherichia coli quinol:fumarate reductase suggest that ligands control domain movement.
Starbird, C.A., Tomasiak, T.M., Singh, P.K., Yankovskaya, V., Maklashina, E., Eisenbach, M., Cecchini, G., Iverson, T.M.(2018) J Struct Biol 202: 100-104
- PubMed: 29158068
- DOI: https://doi.org/10.1016/j.jsb.2017.11.004
- Primary Citation of Related Structures:
6AWF - PubMed Abstract:
Quinol:fumarate reductase (QFR) is an integral membrane protein and a member of the respiratory Complex II superfamily. Although the structure of Escherichia coli QFR was first reported almost twenty years ago, many open questions of catalysis remain. Here we report two new crystal forms of QFR, one grown from the lipidic cubic phase and one grown from dodecyl maltoside micelles. QFR crystals grown from the lipid cubic phase processed as P1, merged to 7.5 Å resolution, and exhibited crystal packing similar to previous crystal forms. Crystals grown from dodecyl maltoside micelles processed as P2 1 , merged to 3.35 Å resolution, and displayed a unique crystal packing. This latter crystal form provides the first view of the E. coli QFR active site without a dicarboxylate ligand. Instead, an unidentified anion binds at a shifted position. In one of the molecules in the asymmetric unit, this is accompanied by rotation of the capping domain of the catalytic subunit. In the other molecule, this is associated with loss of interpretable electron density for this same capping domain. Analysis of the structure suggests that the ligand adjusts the position of the capping domain.
- Graduate Program in Chemical and Physical Biology, Vanderbilt University, Nashville, TN 37232, United States.
Organizational Affiliation: