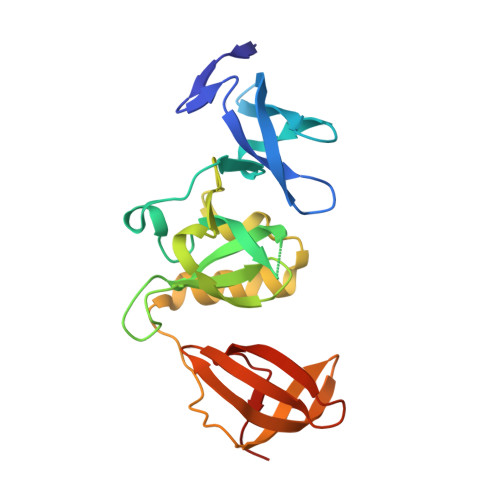
Crystal structure of SETDB1 Tudor domain with aryl triazole fragments
MADER, P., Mendoza-Sanchez, R., IQBAL, A., DONG, A., DOBROVETSKY, E., CORLESS, V.B., LIEW, S.K., TEMPEL, W., SMIL, D., DELA SENA, C.C., KENNEDY, S., DIAZ, D., HOLOWNIA, A., VEDADI, M., BROWN, P.J., SANTHAKUMAR, V., Bountra, C., Edwards, A.M., YUDIN, A.K., Arrowsmith, C.H., Structural Genomics Consortium (SGC)To be published.