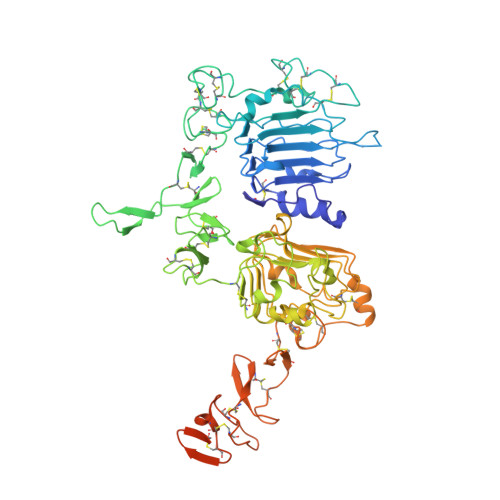
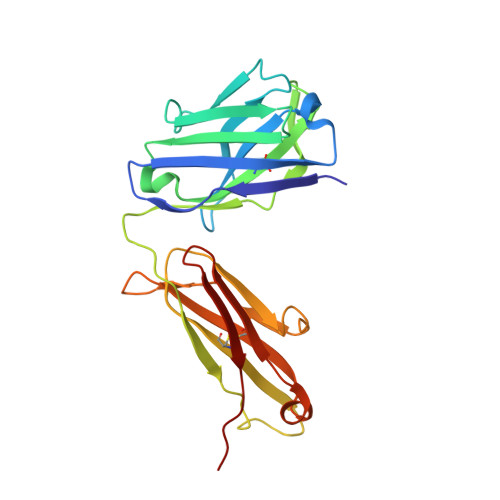
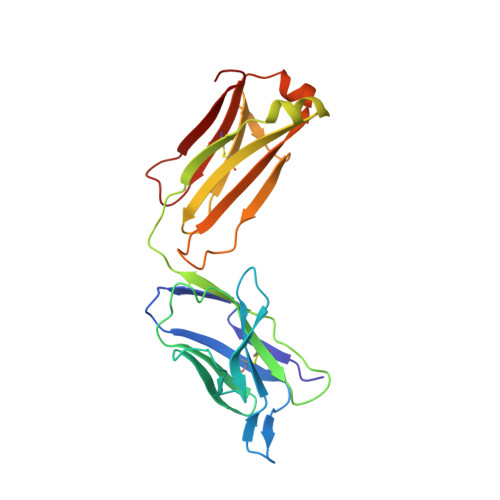
Structural insights into the mechanism of action of a biparatopic anti-HER2 antibody.
Oganesyan, V., Peng, L., Bee, J.S., Li, J., Perry, S.R., Comer, F., Xu, L., Cook, K., Senthil, K., Clarke, L., Rosenthal, K., Gao, C., Damschroder, M., Wu, H., Dall'Acqua, W.(2018) J Biological Chem 293: 8439-8448
- PubMed: 29669810
- DOI: https://doi.org/10.1074/jbc.M117.818013
- Primary Citation of Related Structures:
6ATT - PubMed Abstract:
Pathways of human epidermal growth factor (EGF) receptors are activated upon ligand-dependent or -independent homo- or heterodimerization and their subsequent transphosphorylation. Overexpression of these receptors positively correlates with transphosphorylation rates and increased tumor growth rates. MEDI4276, an anti-human epidermal growth factor receptor 2 (HER2) biparatopic antibody-drug conjugate, has two paratopes within each antibody arm. One, 39S, is aiming at the HER2 site involved in receptor dimerization and the second, single chain fragment (scFv), mimicking trastuzumab. Here we present the cocrystal structure of the 39S Fab-HER2 complex and, along with biophysical and functional assays, determine the corresponding epitope of MEDI4276 and its underlying mechanism of action. Our results reveal that MEDI4276's uniqueness is based first on the ability of its 39S paratope to block HER2 homo- or heterodimerization and second on its ability to cluster the receptors on the surface of receptor-overexpressing cells.
- From the Departments of Antibody Discovery and Protein Engineering, oganesyanv@medimmune.com.
Organizational Affiliation: