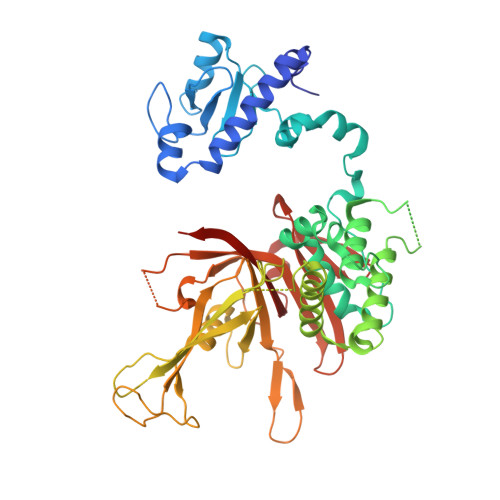
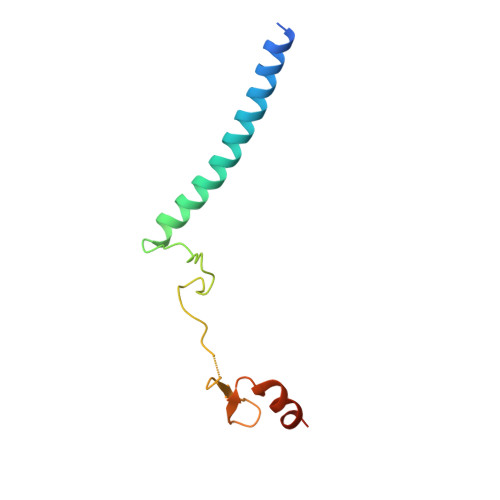
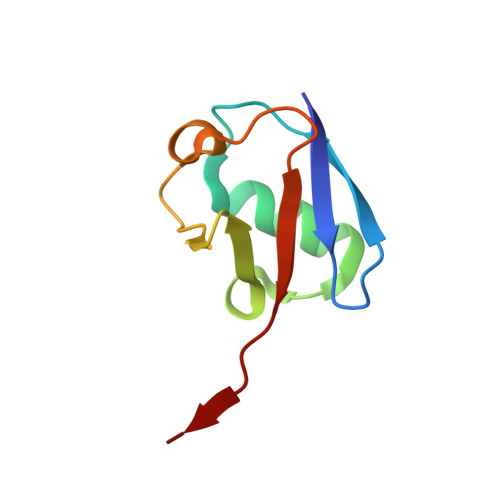
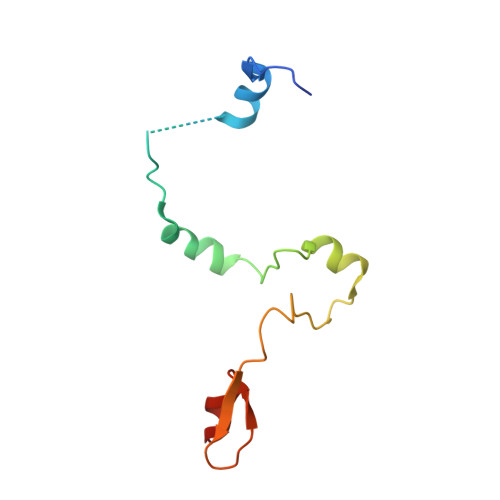
Active site alanine mutations convert deubiquitinases into high-affinity ubiquitin-binding proteins.
Morrow, M.E., Morgan, M.T., Clerici, M., Growkova, K., Yan, M., Komander, D., Sixma, T.K., Simicek, M., Wolberger, C.(2018) EMBO Rep 19
- PubMed: 30150323
- DOI: https://doi.org/10.15252/embr.201745680
- Primary Citation of Related Structures:
6AQR - PubMed Abstract:
A common strategy for exploring the biological roles of deubiquitinating enzymes (DUBs) in different pathways is to study the effects of replacing the wild-type DUB with a catalytically inactive mutant in cells. We report here that a commonly studied DUB mutation, in which the catalytic cysteine is replaced with alanine, can dramatically increase the affinity of some DUBs for ubiquitin. Overexpression of these tight-binding mutants thus has the potential to sequester cellular pools of monoubiquitin and ubiquitin chains. As a result, cells expressing these mutants may display unpredictable dominant negative physiological effects that are not related to loss of DUB activity. The structure of the SAGA DUB module bound to free ubiquitin reveals the structural basis for the 30-fold higher affinity of Ubp8 C146A for ubiquitin. We show that an alternative option, substituting the active site cysteine with arginine, can inactivate DUBs while also decreasing the affinity for ubiquitin.
- Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD, USA.
Organizational Affiliation: