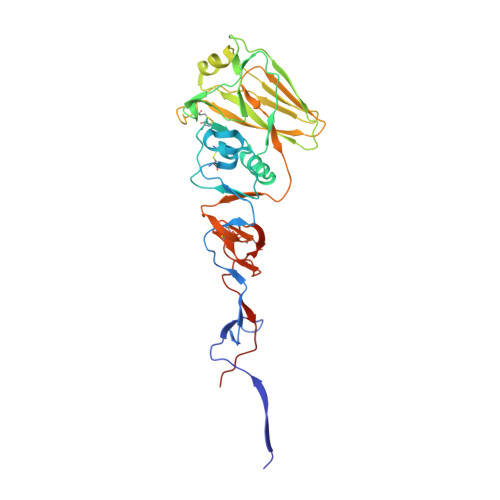
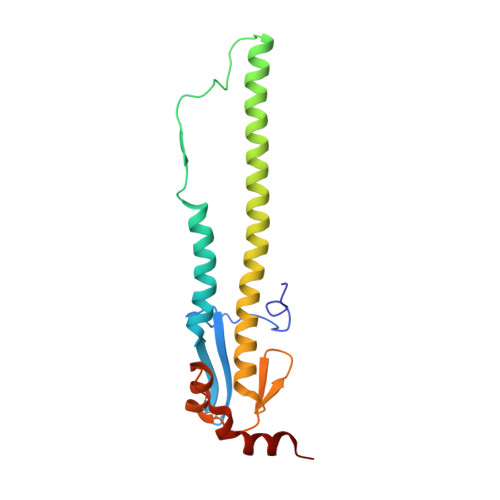
A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine.
Wu, N.C., Zost, S.J., Thompson, A.J., Oyen, D., Nycholat, C.M., McBride, R., Paulson, J.C., Hensley, S.E., Wilson, I.A.(2017) PLoS Pathog 13: e1006682-e1006682
- PubMed: 29059230
- DOI: https://doi.org/10.1371/journal.ppat.1006682
- Primary Citation of Related Structures:
6AOP, 6AOQ, 6AOR, 6AOS, 6AOT, 6AOU, 6AOV - PubMed Abstract:
The effectiveness of the annual influenza vaccine has declined in recent years, especially for the H3N2 component, and is a concern for global public health. A major cause for this lack in effectiveness has been attributed to the egg-based vaccine production process. Substitutions on the hemagglutinin glycoprotein (HA) often arise during virus passaging that change its antigenicity and hence vaccine effectiveness. Here, we characterize the effect of a prevalent substitution, L194P, in egg-passaged H3N2 viruses. X-ray structural analysis reveals that this substitution surprisingly increases the mobility of the 190-helix and neighboring regions in antigenic site B, which forms one side of the receptor binding site (RBS) and is immunodominant in recent human H3N2 viruses. Importantly, the L194P substitution decreases binding and neutralization by an RBS-targeted broadly neutralizing antibody by three orders of magnitude and significantly changes the HA antigenicity as measured by binding of human serum antibodies. The receptor binding mode and specificity are also altered to adapt to avian receptors during egg passaging. Overall, these findings help explain the low effectiveness of the seasonal vaccine against H3N2 viruses, and suggest that alternative approaches should be accelerated for producing influenza vaccines as well as isolating clinical isolates.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, United States of America.
Organizational Affiliation: