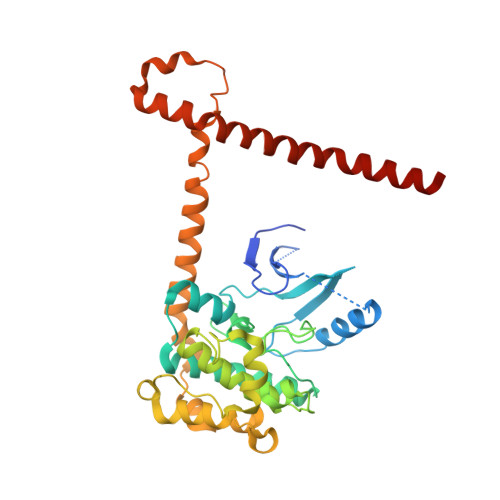
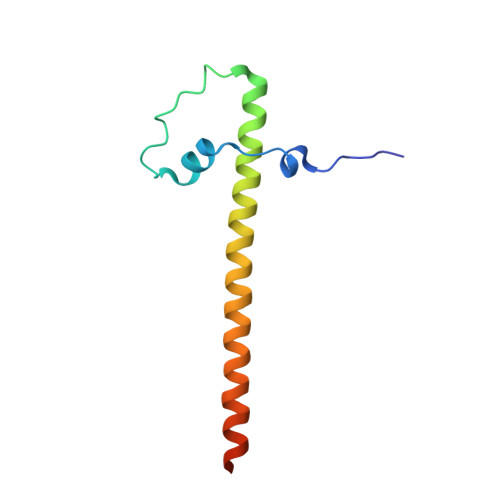
SAV1 promotes Hippo kinase activation through antagonizing the PP2A phosphatase STRIPAK.
Bae, S.J., Ni, L., Osinski, A., Tomchick, D.R., Brautigam, C.A., Luo, X.(2017) Elife 6
- PubMed: 29063833
- DOI: https://doi.org/10.7554/eLife.30278
- Primary Citation of Related Structures:
6AO5, 6AR0, 6AR2 - PubMed Abstract:
The Hippo pathway controls tissue growth and homeostasis through a central MST-LATS kinase cascade. The scaffold protein SAV1 promotes the activation of this kinase cascade, but the molecular mechanisms remain unknown. Here, we discover SAV1-mediated inhibition of the PP2A complex STRIPAK SLMAP as a key mechanism of MST1/2 activation. SLMAP binding to autophosphorylated MST2 linker recruits STRIPAK and promotes PP2A-mediated dephosphorylation of MST2 at the activation loop. Our structural and biochemical studies reveal that SAV1 and MST2 heterodimerize through their SARAH domains. Two SAV1-MST2 heterodimers further dimerize through SAV1 WW domains to form a heterotetramer, in which MST2 undergoes trans-autophosphorylation. SAV1 directly binds to STRIPAK and inhibits its phosphatase activity, protecting MST2 activation-loop phosphorylation. Genetic ablation of SLMAP in human cells leads to spontaneous activation of the Hippo pathway and alleviates the need for SAV1 in Hippo signaling. Thus, SAV1 promotes Hippo activation through counteracting the STRIPAK SLMAP PP2A phosphatase complex.
- Department of Pharmacology, University of Texas Southwestern Medical Center, Dallas, United States.
Organizational Affiliation: