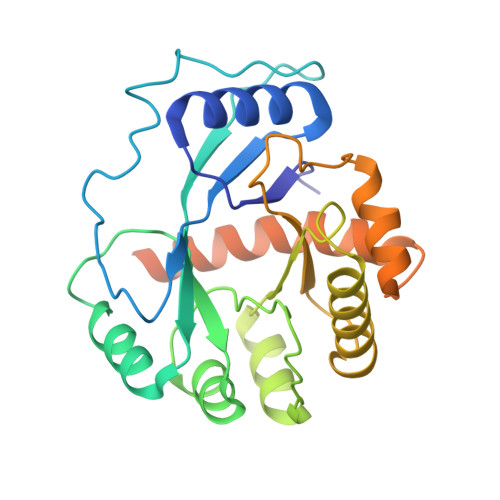
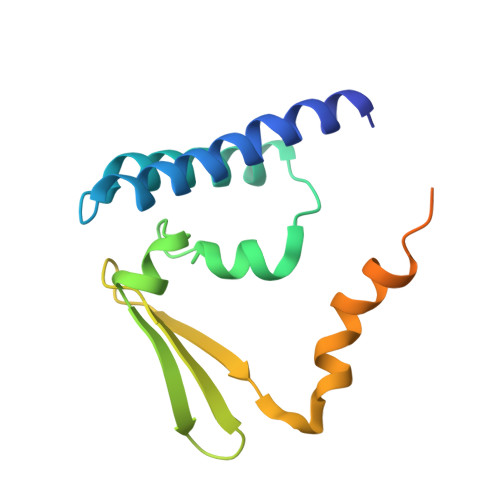
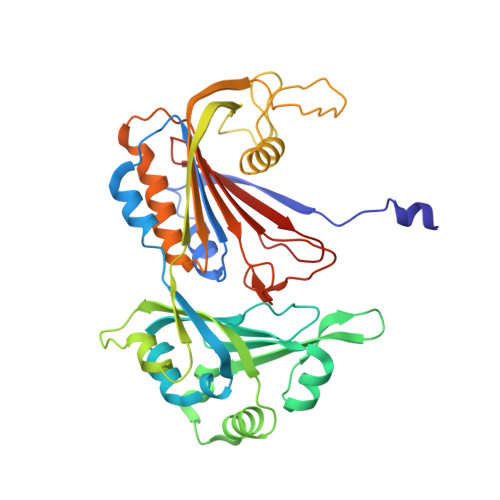
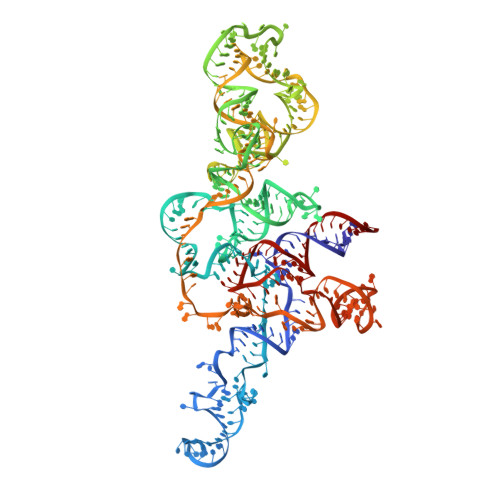
Cryo-EM Structure of the Human Ribonuclease P Holoenzyme.
Wu, J., Niu, S., Tan, M., Huang, C., Li, M., Song, Y., Wang, Q., Chen, J., Shi, S., Lan, P., Lei, M.(2018) Cell 175: 1393-1404.e11
- PubMed: 30454648
- DOI: https://doi.org/10.1016/j.cell.2018.10.003
- Primary Citation of Related Structures:
6AHR, 6AHU, 6AHV - PubMed Abstract:
Ribonuclease (RNase) P is a ubiquitous ribozyme that cleaves the 5' leader from precursor tRNAs. Here, we report cryo-electron microscopy structures of the human nuclear RNase P alone and in complex with tRNA Val . Human RNase P is a large ribonucleoprotein complex that contains 10 protein components and one catalytic RNA. The protein components form an interlocked clamp that stabilizes the RNA in a conformation optimal for substrate binding. Human RNase P recognizes the tRNA using a double-anchor mechanism through both protein-RNA and RNA-RNA interactions. Structural comparison of the apo and tRNA-bound human RNase P reveals that binding of tRNA induces a local conformational change in the catalytic center, transforming the ribozyme into an active state. Our results also provide an evolutionary model depicting how auxiliary RNA elements in bacterial RNase P, essential for substrate binding, and catalysis, were replaced by the much more complex and multifunctional protein components in higher organisms.
- Shanghai Institute of Precision Medicine, Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200125, China.
Organizational Affiliation: