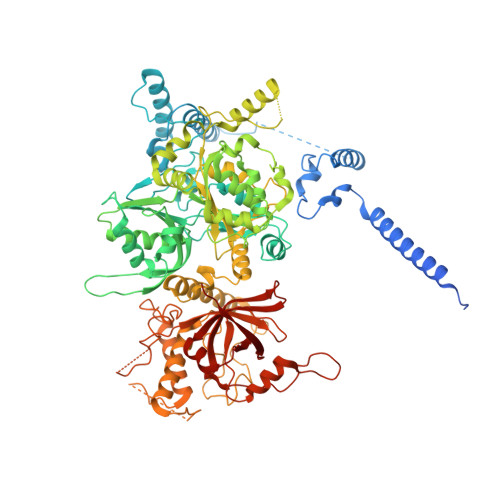
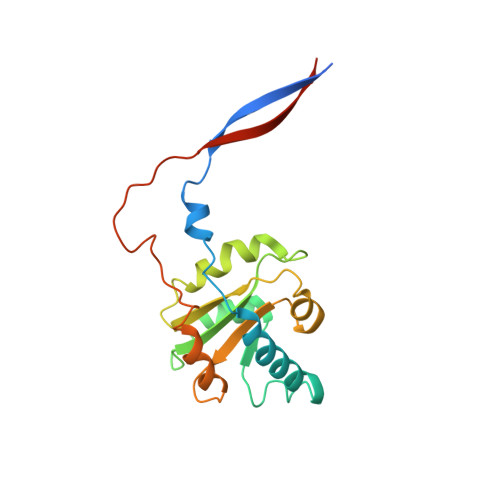
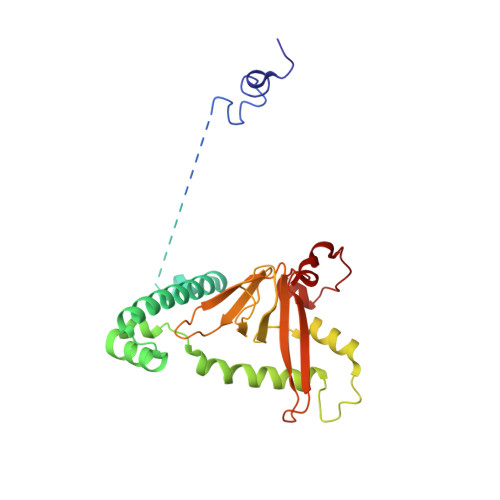
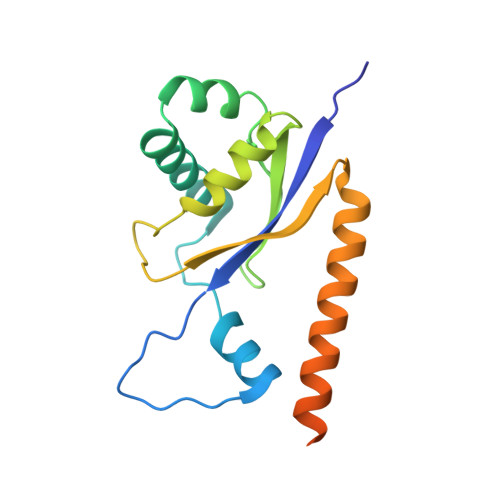
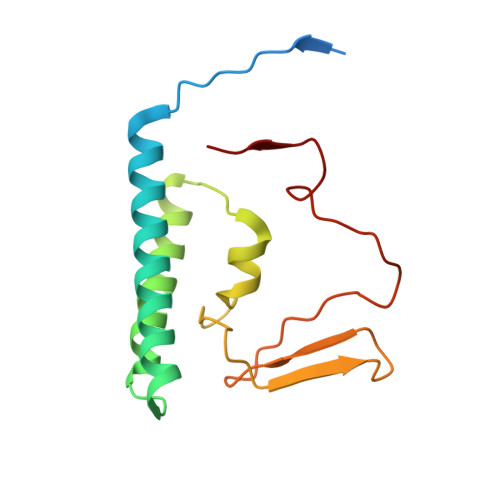
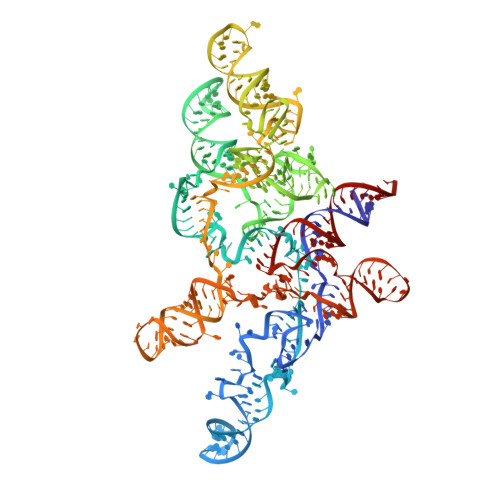
Structural insight into precursor tRNA processing by yeast ribonuclease P.
Lan, P., Tan, M., Zhang, Y., Niu, S., Chen, J., Shi, S., Qiu, S., Wang, X., Peng, X., Cai, G., Cheng, H., Wu, J., Li, G., Lei, M.(2018) Science 362
- PubMed: 30262633
- DOI: https://doi.org/10.1126/science.aat6678
- Primary Citation of Related Structures:
6AGB, 6AH3 - PubMed Abstract:
Ribonuclease P (RNase P) is a universal ribozyme responsible for processing the 5'-leader of pre-transfer RNA (pre-tRNA). Here, we report the 3.5-angstrom cryo-electron microscopy structures of Saccharomyces cerevisiae RNase P alone and in complex with pre-tRNA Phe The protein components form a hook-shaped architecture that wraps around the RNA and stabilizes RNase P into a "measuring device" with two fixed anchors that recognize the L-shaped pre-tRNA. A universally conserved uridine nucleobase and phosphate backbone in the catalytic center together with the scissile phosphate and the O3' leaving group of pre-tRNA jointly coordinate two catalytic magnesium ions. Binding of pre-tRNA induces a conformational change in the catalytic center that is required for catalysis. Moreover, simulation analysis suggests a two-metal-ion S N 2 reaction pathway of pre-tRNA cleavage. These results not only reveal the architecture of yeast RNase P but also provide a molecular basis of how the 5'-leader of pre-tRNA is processed by eukaryotic RNase P.
- Shanghai Institute of Precision Medicine, Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai 200125, China.
Organizational Affiliation: