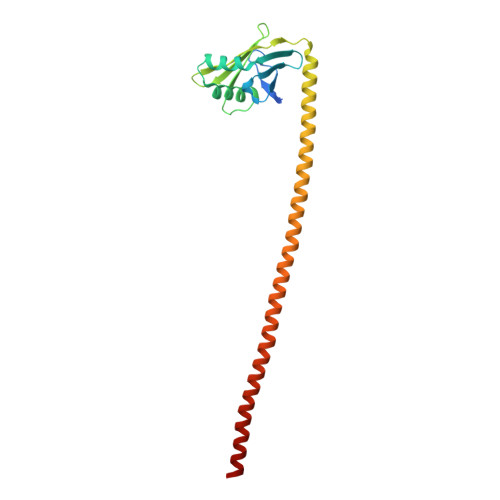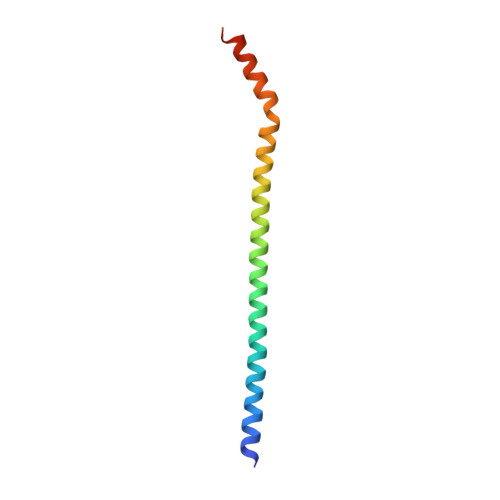The nucleoskeleton protein IFFO1 immobilizes broken DNA and suppresses chromosome translocation during tumorigenesis.
Li, W., Bai, X., Li, J., Zhao, Y., Liu, J., Zhao, H., Liu, L., Ding, M., Wang, Q., Shi, F.Y., Hou, M., Ji, J., Gao, G., Guo, R., Sun, Y., Liu, Y., Xu, D.(2019) Nat Cell Biol 21: 1273-1285
- PubMed: 31548606
- DOI: https://doi.org/10.1038/s41556-019-0388-0
- Primary Citation of Related Structures:
6ABO - PubMed Abstract:
Chromosome translocation is a major cause of the onset and progression of diverse types of cancers. However, the mechanisms underlying this process remain poorly understood. Here, we identified a non-homologous end-joining protein, IFFO1, which structurally forms a heterotetramer with XRCC4. IFFO1 is recruited to the sites of DNA damage by XRCC4 and promotes the repair of DNA double-strand breaks in a parallel pathway with XLF. Interestingly, IFFO1 interacts with lamin A/C, forming an interior nucleoskeleton. Inactivating IFFO1 or its interaction with XRCC4 or lamin A/C leads to increases in both the mobility of broken ends and the frequency of chromosome translocation. Importantly, the destruction of this nucleoskeleton accounts for the elevated frequency of chromosome translocation in many types of cancer cells. Our results reveal that the lamin A/C-IFFO1-constituted nucleoskeleton prevents chromosome translocation by immobilizing broken DNA ends during tumorigenesis.
- State Key Laboratory of Protein and Plant Gene Research, School of Life Sciences, Peking University, Beijing, China.
Organizational Affiliation: