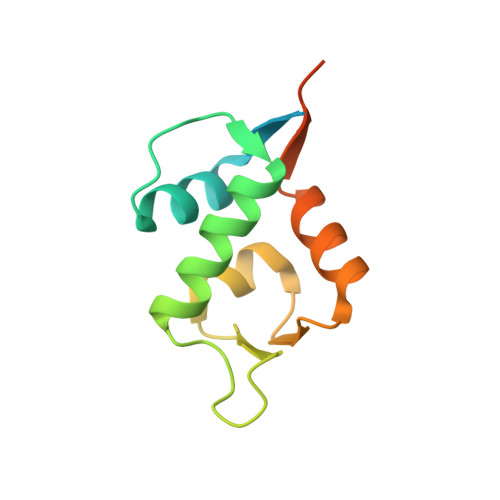
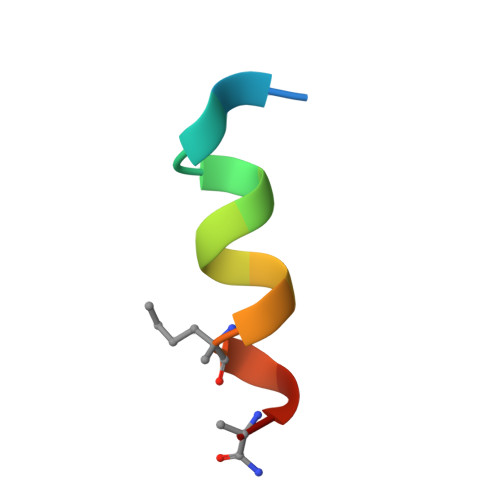
A Model System to Explore Macrocyclic Peptide Structural and Chirality Relationships: Tolerance of helix-breaking residues within the context of a stapled peptide.
Partridge, A.W., Hung, Y.K.K., Juang, Y., Sadruddin, A., Lim, S., Brown, C.J., Ng, S., Thean, D., Ferrer, F., Johannes, C., Yuen, T.Y., Kannan, S., Aronica, P., Tan, Y.S., Mohan, P.R., Verma, C.S., Hichman, J., Chen, S., Wan, H., Lane, D.P., Sawyer, T.K.To be published.