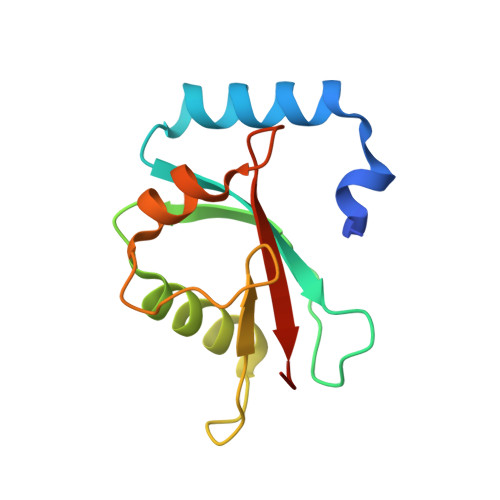
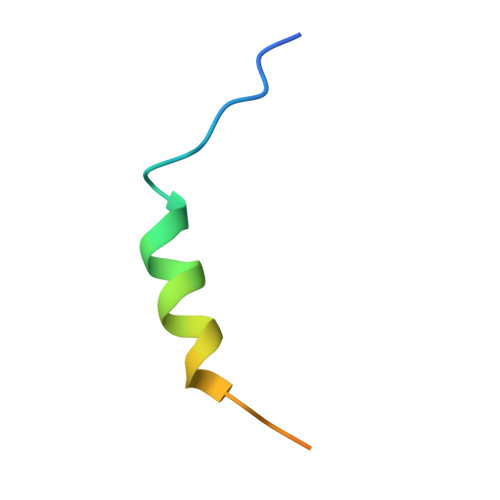
Lipidation-independent vacuolar functions of Atg8 rely on its noncanonical interaction with a vacuole membrane protein
Liu, X.M., Yamasaki, A., Du, X.M., Coffman, V.C., Ohsumi, Y., Nakatogawa, H., Wu, J.Q., Noda, N.N., Du, L.L.(2018) Elife 7
- PubMed: 30451685
- DOI: https://doi.org/10.7554/eLife.41237
- Primary Citation of Related Structures:
6AAF, 6AAG - PubMed Abstract:
The ubiquitin-like protein Atg8, in its lipidated form, plays central roles in autophagy. Yet, remarkably, Atg8 also carries out lipidation-independent functions in non-autophagic processes. How Atg8 performs its moonlighting roles is unclear. Here we report that in the fission yeast Schizosaccharomyces pombe and the budding yeast Saccharomyces cerevisiae , the lipidation-independent roles of Atg8 in maintaining normal morphology and functions of the vacuole require its interaction with a vacuole membrane protein Hfl1 (homolog of human TMEM184 proteins). Crystal structures revealed that the Atg8-Hfl1 interaction is not mediated by the typical Atg8-family-interacting motif (AIM) that forms an intermolecular β-sheet with Atg8. Instead, the Atg8-binding regions in Hfl1 proteins adopt a helical conformation, thus representing a new type of AIMs (termed helical AIMs here). These results deepen our understanding of both the functional versatility of Atg8 and the mechanistic diversity of Atg8 binding.
- National Institute of Biological Sciences, Beijing, China.
Organizational Affiliation: