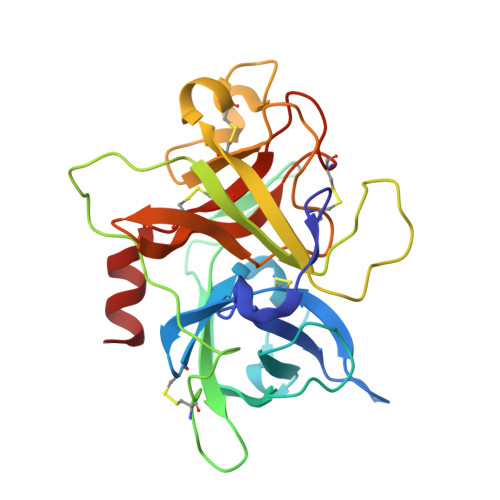
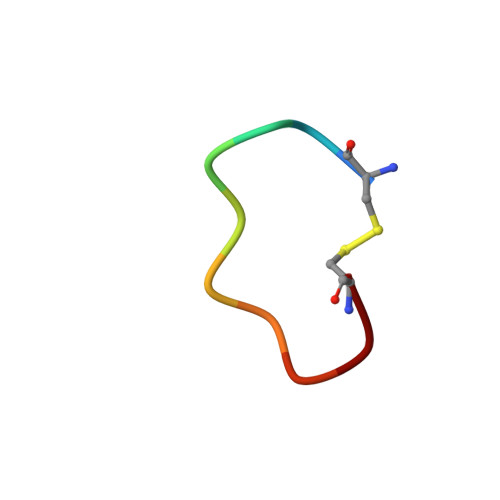
Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
Wang, D., Yang, Y., Jiang, L., Wang, Y., Li, J., Andreasen, P.A., Chen, Z., Huang, M., Xu, P.(2019) J Med Chem 62: 2172-2183
- PubMed: 30707839
- DOI: https://doi.org/10.1021/acs.jmedchem.8b01908
- Primary Citation of Related Structures:
6A8G, 6A8N - PubMed Abstract:
Urokinase-type plasminogen activator (uPA) is a diagnostic marker for breast and prostate cancers recommended by American Society for Clinical Oncology and German Breast Cancer Society. Inhibition of uPA was proposed as an efficient strategy for cancer treatments. In this study, we report peptide-based uPA inhibitors with high potency and specificity comparable to monoclonal antibodies. We revealed the binding and inhibitory mechanisms by combining crystallography, molecular dynamic simulation, and other biophysical and biochemical approaches. Besides, we showed that our peptides efficiently inhibited the invasion of cancer cells via intervening with the processes of the degradation of extracellular matrices. Furthermore, our peptides significantly suppressed the tumor growth and the cancer metastases in tumor-bearing mice. This study demonstrates that these uPA peptides are highly potent anticancer agents and reveals the mechanistic insights of these uPA inhibitors, which can be useful for developing other serine protease inhibitors.
- State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter , Chinese Academy of Sciences , 155 West Yangqiao Road , Fuzhou , Fujian 350002 , China.
Organizational Affiliation: