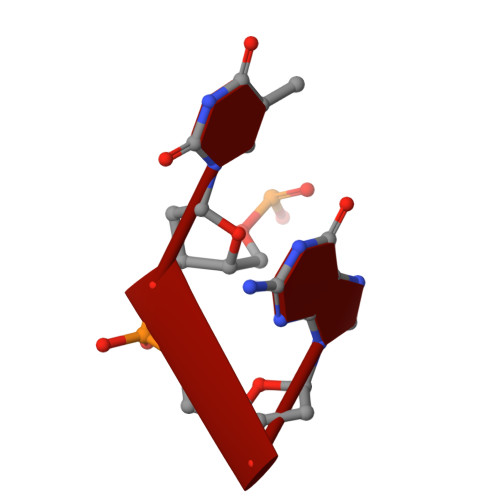
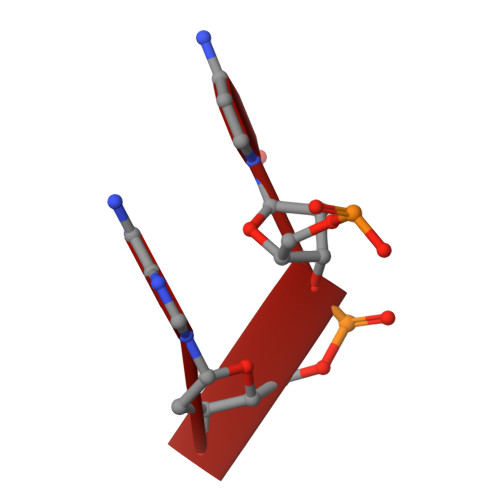
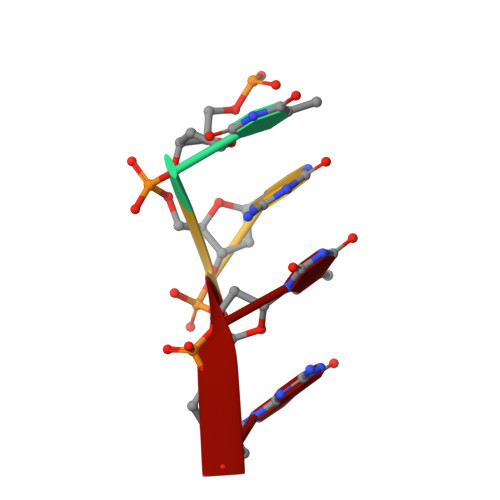
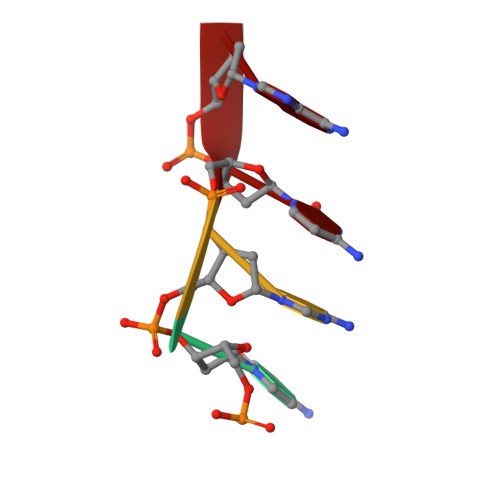
Crystal structures of disordered Z-type helices.
Karthik, S., Mandal, P.K., Thirugnanasambandam, A., Gautham, N.(2019) Nucleosides Nucleotides Nucleic Acids 38: 279-293
- PubMed: 30588873
- DOI: https://doi.org/10.1080/15257770.2018.1517883
- Primary Citation of Related Structures:
6A1O, 6A1Q - PubMed Abstract:
The X-ray crystal structures of a decamer sequence d(CGCGTACGCG) 2 and a tetradecamer sequence d(CGCGCGTACGCGCG) 2 are presented here. Both sequences are alternating pyrimidine-purine repeat sequences and they form disordered, pseudo-continuous left handed Z-type helices. They demonstrate interesting variants of the 'bundles of columns of helices' mode of packing.
- a Centre of Advanced Study in Crystallography and Biophysics , University of Madras, Guindy Campus , Chennai , Tamil Nadu , India.
Organizational Affiliation: