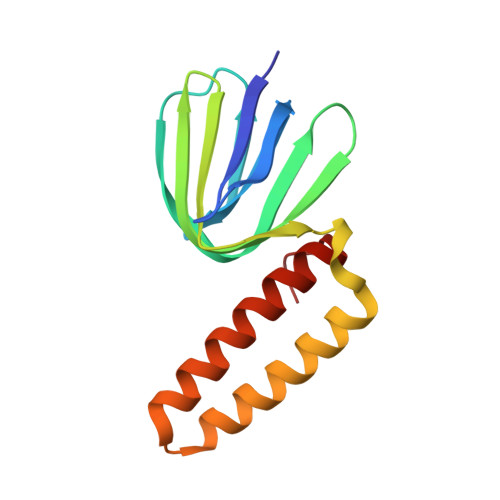
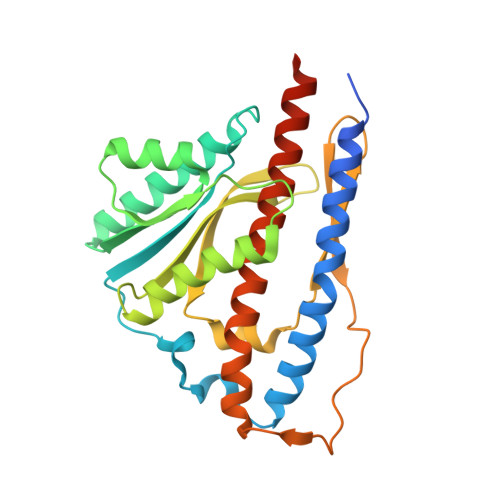
Structure of the gamma-epsilon complex of cyanobacterial F1-ATPase reveals a suppression mechanism of the gamma subunit on ATP hydrolysis in phototrophs.
Murakami, S., Kondo, K., Katayama, S., Hara, S., Sunamura, E.I., Yamashita, E., Groth, G., Hisabori, T.(2018) Biochem J 475: 2925-2939
- PubMed: 30054433
- DOI: https://doi.org/10.1042/BCJ20180481
- Primary Citation of Related Structures:
5ZWL - PubMed Abstract:
F 1 -ATPase forms the membrane-associated segment of F 0 F 1 -ATP synthase - the fundamental enzyme complex in cellular bioenergetics for ATP hydrolysis and synthesis. Here, we report a crystal structure of the central F 1 subcomplex, consisting of the rotary shaft γ subunit and the inhibitory ε subunit, from the photosynthetic cyanobacterium Thermosynechococcus elongatus BP-1, at 1.98 Å resolution. In contrast with their homologous bacterial and mitochondrial counterparts, the γ subunits of photosynthetic organisms harbour a unique insertion of 35-40 amino acids. Our structural data reveal that this region forms a β-hairpin structure along the central stalk. We identified numerous critical hydrogen bonds and electrostatic interactions between residues in the hairpin and the rest of the γ subunit. To elaborate the critical function of this β-hairpin in inhibiting ATP hydrolysis, the corresponding domain was deleted in the cyanobacterial F 1 subcomplex. Biochemical analyses of the corresponding α 3 β 3 γ complex confirm that the clinch of the hairpin structure plays a critical role and accounts for a significant interaction in the α 3 β 3 complex to induce ADP inhibition during ATP hydrolysis. In addition, we found that truncating the β-hairpin insertion structure resulted in a marked impairment of the interaction with the ε subunit, which binds to the opposite side of the γ subunit from the β-hairpin structure. Combined with structural analyses, our work provides experimental evidence supporting the molecular principle of how the insertion region of the γ subunit suppresses F 1 rotation during ATP hydrolysis.
- Department of Life Science, Tokyo Institute of Technology, Nagatsuta, Midori-ku, Yokohama 226-8501, Japan.
Organizational Affiliation: