Structural and biochemical characterization of a multidomain alginate lyase reveals a novel role of CBM32 in CAZymes
Lyu, Q., Zhang, K., Zhu, Q., Li, Z., Liu, Y., Fitzek, E., Yohe, T., Zhao, L., Li, W., Liu, T., Yin, Y., Liu, W.(2018) Biochim Biophys Acta 1862: 1862-1869
- PubMed: 29864445
- DOI: https://doi.org/10.1016/j.bbagen.2018.05.024
- Primary Citation of Related Structures:
5ZU5, 5ZU6 - PubMed Abstract:
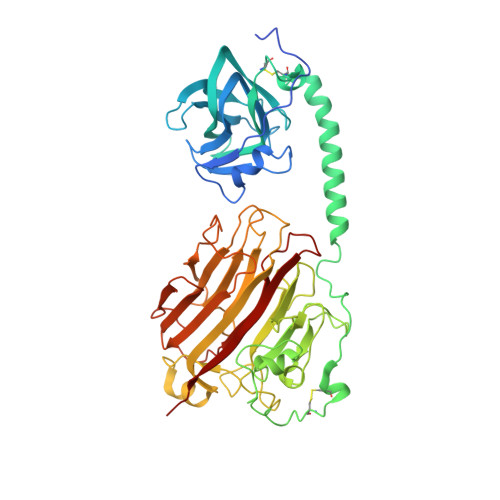
Noncatalytic carbohydrate binding modules (CBMs) have been demonstrated to play various roles with cognate catalytic domains. However, for polysaccharide lyases (PLs), the roles of CBMs remain mostly unknown. AlyB is a multidomain alginate lyase that contains CBM32 and a PL7 catalytic domain. The AlyB structure determined herein reveals a noncanonical alpha helix linker between CBM32 and the catalytic domain. More interestingly, CBM32 and the linker does not significantly enhance the catalytic activity but rather specifies that trisaccharides are predominant in the degradation products. Detailed mutagenesis, biochemical and cocrystallization analyses show "weak but important" CBM32 interactions with alginate oligosaccharides. In combination with molecular modeling, we propose that the CBM32 domain serves as a "pivot point" during the trisaccharide release process. Collectively, this work demonstrates a novel role of CBMs in the activity of the appended PL domain and provides a new avenue for the well-defined generation of alginate oligosaccharides by taking advantage of associated CBMs.
- MOE Key Laboratory of Marine Genetics and Breeding, College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China; Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao 266235, China.
Organizational Affiliation: