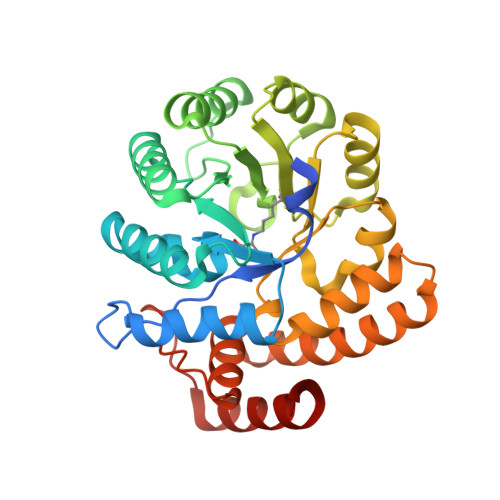
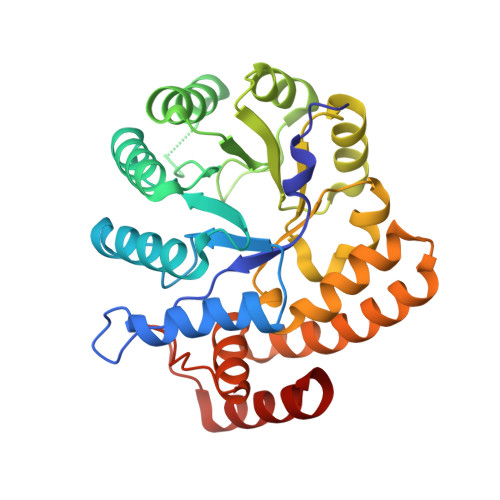
Crystal structures and kinetics of N-acetylneuraminate lyase from Fusobacterium nucleatum
Kumar, J.P., Rao, H., Nayak, V., Ramaswamy, S.(2018) Acta Crystallogr F Struct Biol Commun 74: 725-732
- PubMed: 30387778
- DOI: https://doi.org/10.1107/S2053230X18012992
- Primary Citation of Related Structures:
5ZJM, 5ZKA - PubMed Abstract:
N-Acetyl-D-neuraminic acid lyase (NanA) catalyzes the breakdown of sialic acid (Neu5Ac) to N-acetyl-D-mannosamine (ManNAc) and pyruvate. NanA plays a key role in Neu5Ac catabolism in many pathogenic and bacterial commensals where sialic acid is available as a carbon and nitrogen source. Several pathogens or commensals decorate their surfaces with sialic acids as a strategy to escape host innate immunity. Catabolism of sialic acid is key to a range of host-pathogen interactions. In this study, atomic resolution structures of NanA from Fusobacterium nucleatum (FnNanA) in ligand-free and ligand-bound forms are reported at 2.32 and 1.76 Å resolution, respectively. F. nucleatum is a Gram-negative pathogen that causes gingival and periodontal diseases in human hosts. Like other bacterial N-acetylneuraminate lyases, FnNanA also shares the triosephosphate isomerase (TIM)-barrel fold. As observed in other homologous enzymes, FnNanA forms a tetramer. In order to characterize the structure-function relationship, the steady-state kinetic parameters of the enzyme are also reported.
- Technologies for the Advancement of Science, Institute for Stem Cell Biology and Regenerative Medicine, NCBS, GKVK Campus, Bangalore, Karnataka 560 065, India.
Organizational Affiliation: